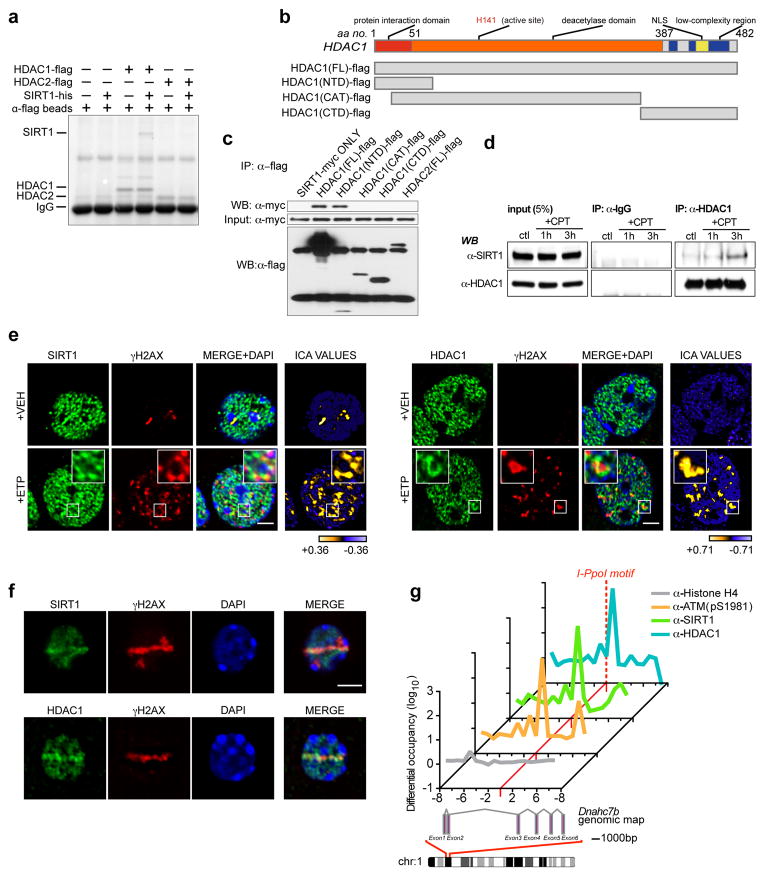
Figure 2. SIRT1 and HDAC1 physically interact and localize to DSB sites in neurons.
a, Recombinant SIRT1-his was incubated with either HDAC1-flag or HDAC2-flag, following which HDAC1 and HDAC2 were precipitated with anti-flag conjugated agarose beads, and their ability to retain SIRT1 was assessed. b, Diagram illustrating HDAC1 fragment constructs for interaction mapping. FL – full-length; CTD – carboxy-terminal domain; CAT – catalytic domain; NTD – amino-terminal domain. c, The indicated flag-tagged fragments were expressed together with SIRT1-myc, and the flag-tagged proteins were immunoprecipitated and blotted with antibodies against myc. d, HT22 cells were treated with camptothecin (CPT; 1μM), precipitated with antibodies against HDAC1, and blotted with antibodies against SIRT1. e, Etoposide-treated primary neurons treated were fixed and stained with antibodies against either SIRT1 or HDAC1, and γH2AX. Right-most panel illustrates Intensity Correlation Analysis (ICA). Pixels from input channel co-varying positively with corresponding signal from γH2AX channel are indicated in yellow, while negatively co-varying pixels are indicated in blue. Scale bar = 3μm. f, Primary neurons were subjected to sub-nuclear, laser-generated DNA lesioning and stained with antibodies against either SIRT1 or HDAC1, and γH2AX. Scale bar = 3μm. g, The rare-cutting homing endonuclease, I-PpoI, was used to generate DNA DSBs at defined genetic loci in mouse cortical neurons. Recruitment of the indicated proteins at a unique cleavage site located between exons 2 and 3 in the Dnahc7b gene was then assessed by ChIP. Primers were designed at regular 1kb intervals, spanning 10kb both 3’ and 5’ to the I-PpoI consensus site (red dotted line).