Abstract
The present study aimed to explore the molecular mechanisms underlying the development of thoracic spinal cord injury (SCI). The gene expression profile of GSE20907, which included 12 thoracic non-injured spinal cord control samples and 12 thoracic transected spinal cord samples at different stages of SCI, was obtained from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were identified using the limma package in R/Bioconductor. DEG-associated pathways were analyzed using the Kyoto encyclopedia of genes and genomes database. A protein-protein interaction (PPI) network was constructed and transcription factors (TFs) were predicted using cytoscape. Compared with the control samples, there were 1,942, 396, 188 and 396 DEGs identified at day 3 (d3), week 1 (wk1), wk2 and month 1 (m1), respectively. Cluster analysis indicated that the DEGs at m1 were similar to those in the control group. Downregulated DEGs were enriched in nervous system disease pathways, such as Parkinson's disease. Upregulated DEGs were enriched in immune response-associated pathways, such as Fc γ R-mediated phagocytosis at early stages (d3 and wk1). Upregulated DEGs were enriched in pathways associated with cancer and pyrimidine metabolism at wk2 and m1, respectively. In the PPI network, nodes including RAC2, CD4, STAT3 and JUN were identified. Furthermore, ATF3, JUN and EGR1 were identified as TFs associated with SCI. In conclusion, the results of the present study showed that the number of DEGs decreased in a time-dependent manner following SCI. OLIG1, ATF3 and JUN may represent SCI regeneration-associated genes. Immune-associated inflammation was shown to be important in SCI, and SCI exhibits causal associations with other diseases, including cardiovascular disease and cancers. The present study provided novel insight into the molecular mechanisms of SCI regeneration, which may aid in the development of strategies to enhance recovery following SCI.
Keywords: spinal cord injury, differentially expressed gene, pathway, protein-protein interaction, transcription factors
Introduction
Spinal cord injury (SCI) is predominantly caused by accidents associated with falls, vehicle collisions and sport. Every year there are ~12,000 novel cases of SCI reported in the USA (1) and 60,000 novel cases in China, which represents the greatest incidence world-wide (2). SCI may lead to paraplegia or quadriplegia and patients may be permanently physically disabled (3,4). Patients with SCI are often confined to a wheelchair (5). Recent imaging studies have been developed for predicting the outcomes for patients with SCI (4). Further investigations into the mechanisms for regeneration and functional restoration of patients with SCI are required. Recent advances in neuroscience research have provided novel insight into the rehabilitation of patients with SCI. A number of rehabilitative, cellular and molecular therapies have been tested, using animal models and clinical trials (3,6).
SCI is a form of central nervous system (CNS) trauma. Regenerative mechanisms of the CNS are typically suppressed in response to a number of extrinsic and intrinsic factorsincluding Nogo, glial scars and chondroitin sulfate proteoglycan activity (7). Phospholipase A2 (PLA2) mediates multiple injury mechanisms following SCI and may represent a novel and efficient strategy for inhibiting a number of injury pathways that occur following SCI (8). Inflammation is one of the consequences of CNS trauma (9). Histone H3K27me3 demethylation of PLA2 may regulate acute inflammatory responses and improve the blood-spinal cord barrier following SCI (10). Immune cells, including macrophages and B- and T cells, may protect and repair the injured CNS, and the latter two are capable of secreting the bio-active form of brain-derived neurotrophic factor (11). Previous studies have demonstrated that the CNS is associated with other diseases, including hypertension, cardiovascular diseases (12,13) and cancers (14,15). However, the underlying mechanisms of SCI development and regeneration have remained to be fully elucidated.
Recent bioinformatic analyses have explored the genetic processes and molecular mechanisms underlying SCI. Siebert et al (16) analyzed the cellular response of thoracic propriospinal neurons and the regenerative ability following low thoracic complete SCI using the gene expression profile of GSE20907. Lai et al (17) identified a number of SCI-associated pathways, including cell cycle, immune response and olfactory transduction. Jin et al (18) found that cell cycle and immune system-associated pathways, as well as oxidative phosphorylation and CNS disease signaling pathways are important in the development of SCI. However, Lai et al (17) demonstrated that the identification of SCI-associated genes is inconsistent due to the different criteria used for analyzing differentially expressed genes (DEGs). Furthermore, changes in gene expression over time have not been investigated.
Therefore, using the expression profile GSE20907, the present study analyzed time-dependent changes of SCI-associated DEGs with a cutoff criterion of P<0.01 and Fold-changes of gene expression (log2 FC) ≥1. In addition, the sub-pathways in which the DEGs were enriched were identified. Protein-protein interaction (PPI) network construction and transcription factor (TF) annotation were performed in order to explore the 'hub' nodes (highly connected nodes with a large degree) and TFs at various time-points following SCI. The results of the present study provided novel insight into the mechanisms underlying SCI.
Materials and methods
Microarray data
The expression profile GSE20907 based on the Affymetrix Rat Gene 1.0 ST Array (GPL6247; Affymetrix, Inc., Santa Clara, CA, USA) platform was obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/; accessed June 16, 2014). Data included 12 thoracic non-injured spinal cord control samples (Ctrl) and 12 thoracic transected spinal cord samples at 3 days (d3, n=4), 1 week (wk1, n=4), 2 weeks (wk2, n=2) and 1 month (m1, n=2) post-lesion.
Data processing
Expression profile chip data were processed using the affy package (19) in R/Bioconductor, version 2.14.1 (20) (http://www.bioconductor.org/). Data were subjected to background correction, normalization, probe summary and log2 logarithmic transformation using the robust multi-array average (RMA Express; version 1.0; http://rmaexpress.bmbolstad.com) method (21). When several probes were found to project to one gene, the average was used to represent the expression levels of this gene. There were 27,342 probes in the raw data and 15,594 genes remained following data processing.
Identification of DEGs between SCI and controls at four time-points
GSE20907 data included one Ctrl group and four experimental groups at different time-points (d3, wk1, wk2 and m1). Data were divided into four paired groups: d3-Ctrl, wk1-Ctrl, wk2-Ctrl and m1-Ctrl. The Limma package (22) in R/Bioconductor was used to analyze the DEGs in each experimental group. |log2FC| and P-values from Student's t-test were used to select the DEGs. A P-value <0.01 and |log2FC|≥1 were set as the cutoff criteria.
Cluster analysis of DEGs
In order to analyze the changes of DEG expression at the four time-points, the gplots package (23) in R/Bioconductor was used to construct a cluster heatmap of DEGs. Mean expression values of DEGs for the different time-point samples and controls were used to form the expression matrix.
Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis of DEGs
The database for annotation, visualization and integrated discovery (DAVID; version 6.7) provides a comprehensive set of functional annotation tools (24). In order to identify DEG functions, overregulated KEGG (version 59) categories in pathways were identified using DAVID (25,26). DAVID was used to identify DEGs associated pathways by calculating the hyper-geometric test P-values (27). P<0.01 was set as the cut-off criterion.
Construction of the PPI network
The search tool for the retrieval of interacting genes (STRING; version 9.0) database (27) was used to annotate functional interactions between the DEGs encoding proteins. Cytoscape, version 2.6.3 (28) was then used to construct the PPI networks for the DEGs at different stages post-SCI.
TF annotation
Based on the rat TFs database TRANSFAC version 6.0 (http://www.gene-regulation.com) (29), TFs were annotated among DEGs. Using the TF annotation and PPI network information, the differences and similarities between TFs at the four time-points as well as the degrees of TFs in the PPI network were analyzed.
Results
DEGs between the SCI and Ctrl samples at four time-points
According to the gene expression profiles, 1,942, 396, 188 and 193 DEGs were identified at d3, wk1, wk2 and m1, respectively (Table I). The number of DEGs decreased in a time-dependent manner. Upregulated and downregulated DEGs are summarized in Table I. There was a greater number of upregulated DEGs than that of downregulated DEGs at the four time-points.
Table I.
DEG counts (n) at four time-points following spinal cord injury in rats.
| DEGs | Upregulated genes | Downregulated genes | |
|---|---|---|---|
| d3-Ctrl | 1,942 | 1,038 | 904 |
| wk1-Ctrl | 396 | 204 | 192 |
| wk2-Ctrl | 188 | 146 | 42 |
| m1-Ctrl | 193 | 154 | 39 |
DEGs, differentially expressed genes; d3, day 3; wk1, week 1; wk2, week 2; m1, month 1.
Cluster analysis of the DEGs
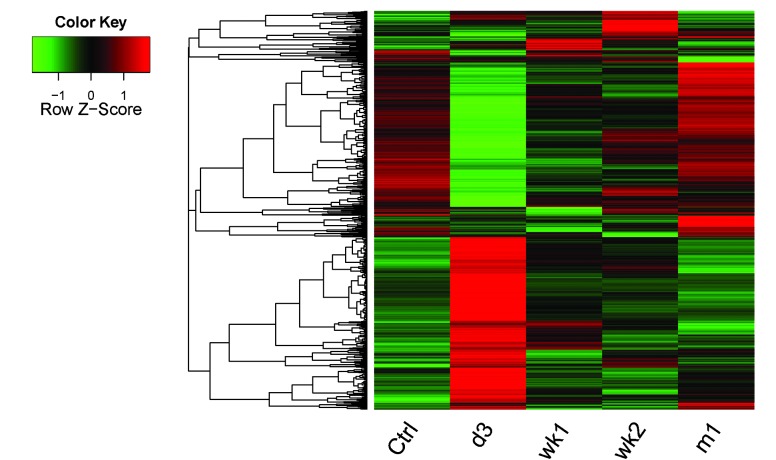
In order to explore the changes of the DEG expression levels at the four time-points following SCI, a cluster analysis was performed. A cluster heat map of the four experimental groups compared with the Ctrl group is shown in Fig. 1. DEG expression levels of d3 samples were markedly different from those of the Ctrl samples. DEG expression levels of m1 samples were similar to those in the Ctrl group.
Figure 1.
Cluster heat map of differentially expressed genes. Relative levels of gene expression are represented using a color scale: Green represents downregulated genes and red represents upregulated gene levels. d3, day 3; wk1, week 1; wk2, week 2; m1, month 1; Ctrl, control.
KEGG pathway enrichment analysis
The KEGG pathways of the significantly upregulated and downregulated genes are summarized in Table II. Results demonstrated that the significantly enriched KEGG pathways of the downregulated genes were relatively similar between d3, wk1 and wk2, which were predominantly associated with pathways of neurological diseases, including Parkinson's disease, oxidative phosphorylation, Huntington's disease and Alzheimer's disease. At d3 and wk1, the upregulated genes were enriched in immune response-associated pathways, including Fc γ R-mediated phagocytosis, lysosome, leukocyte transendothelial migration, B-cell receptor signaling pathway, complement and coagulation cascades, systemic lupus erythematosus and natural killer cell-mediated cytotoxicity. At wk2 and m1, upregulated genes were enriched in pathways associated with cancer and pyrimidine metabolism, respectively. Overall, DEGs were predominantly associated with pathways of immune and nervous system-associated diseases.
Table II.
KEGG pathways of significantly up- and downregulated genes at four time-points following spinal cord injury in rats.
| Contrast group | KEGG pathway | Gene count (n) | P-value |
|---|---|---|---|
| d3-Ctrl | |||
| Upregulated genes | rno04666: Fc gamma R-mediated phagocytosis | 24 | 5.25×10−8 |
| rno04142: Lysosome | 28 | 6.27×10−8 | |
| rno04670: Leukocyte transendothelial migration | 27 | 1.72×10−7 | |
| rno04810: Regulation of actin cytoskeleton | 37 | 1.10×10−6 | |
| rno04662: B cell receptor signaling pathway | 20 | 1.31×10−6 | |
| Downregulated genes | rno05012: Parkinson's disease | 55 | 1.13×10−34 |
| rno00190: Oxidative phosphorylation | 54 | 1.79×10−34 | |
| rno05016: Huntington's disease | 54 | 2.27×10−26 | |
| rno05010: Alzheimer's disease | 54 | 1.56×10−25 | |
| wk1-Ctrl | |||
| Upregulated genes | rno04610: Complement and coagulation cascades | 9 | 4.69×10−06 |
| rno05322: Systemic lupus erythematosus | 8 | 2.32×10−04 | |
| rno04650: Natural killer cell mediated cytotoxicity | 7 | 0.0026 | |
| rno04666: Fc gamma R-mediated phagocytosis | 6 | 0.0074 | |
| Downregulated genes | rno05012: Parkinson's disease | 14 | 9.93×10−10 |
| rno00190: Oxidative phosphorylation | 13 | 7.99×10−9 | |
| rno05016: Huntington's disease | 13 | 3.11×10−7 | |
| rno05010: Alzheimer's disease | 13 | 4.61×10−7 | |
| rno00100: Steroid biosynthesis | 6 | 5.20×10−7 | |
| rno00900: Terpenoid backbone biosynthesis | 5 | 9.02×10−6 | |
| wk2-Ctrl | |||
| Upregulated genes | rno05200: Pathways in cancer | 8 | 0.0035 |
| Downregulated genes | rno05012: Parkinson's disease | 6 | 2.41×10−5 |
| rno05016: Huntington's disease | 6 | 9.79×10−5 | |
| rno04260: Cardiac muscle contraction | 4 | 0.0010 | |
| rno00190: Oxidative phosphorylation | 4 | 0.0053 | |
| m1-Ctrl | |||
| Upregulated genes | rno00240: Pyrimidine metabolism | 6 | 7.87×10−4 |
Contrast group represents the comparison of the control group and the four experimental groups; gene counts are the numbers of genes that are enriched in the pathway. d3, day 3; wk1, week 1; wk2, week 2; m1, month 1; Ctrl, control; KEGG, Kyoto encyclopedia of genes and genomes.
PPI network
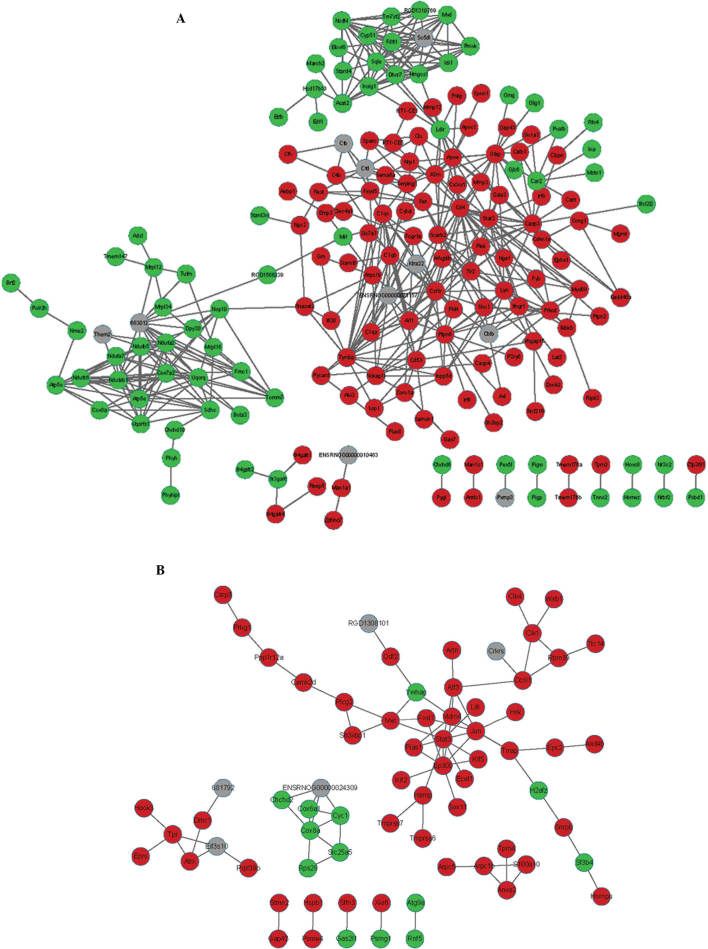
The PPI network of d3 consisted of 1,524 protein nodes and 10,390 pairs of mutual protein associations (data not shown). The PPI network of the DEGs based on three time-points wk1, wk2 and m1 are demonstrated in Figs. 2 and 3. The PPI network of wk1 consisted of 184 protein nodes and 475 protein pairs. THe PPI network of wk2 consisted of 71 protein nodes and 82 protein pairs. The PPI network of m1 consisted of 49 protein nodes and 43 protein pairs.
Figure 2.
Protein-protein interaction networks based on the DEGs. Undirected network at (A) one week and (B) two weeks post-lesion. Green nodes represent proteins coded by the downregulated genes, red nodes represent proteins coded by the upregulated genes and grey nodes represent proteins that are not coded by DEGs. DEG, differentially expressed gene.
Figure 3.
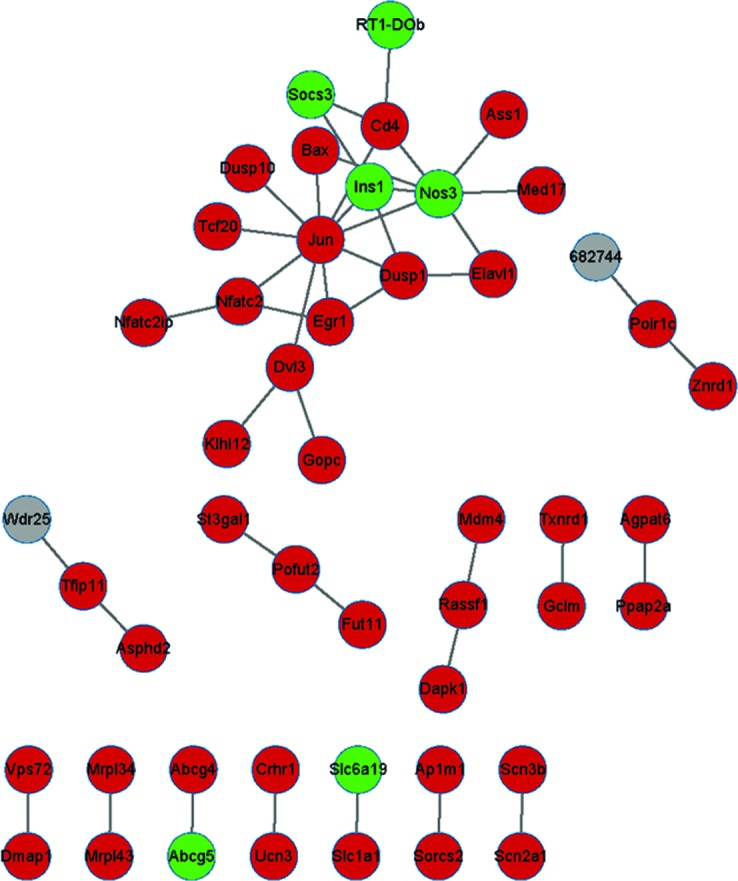
Protein-protein interaction network based on the DEGs. Undirected network at one month post-lesion. Green nodes represent proteins coded by the downregulated genes, red nodes represent proteins coded by the upregulated genes and grey nodes represent proteins that are not coded by DEGs. DEG, differentially expressed gene.
The top 10 hub node genes exhibiting enhanced expression at the four time-points are summarized in Table III. Hub node genes in the PPI network were differentially expressed at the four time-points. STAT3 was the hub gene at wk1 and wk2, JUN was the hub gene at wk2 and m1 and CD4 was the hub gene at d3, wk1 and m1, while RAC2 was the hub gene at d3.
Table III.
Top 10 upregulated hub genes at four time-points following spinal cord injury in rats.
| d3
|
wk1
|
wk2
|
m1
|
||||
|---|---|---|---|---|---|---|---|
| Gene | Degree | Gene | Degree | Gene | Degree | Gene | Degree |
| Rac2 | 130 | Cd4 | 22 | Stat3 | 9 | Jun | 10 |
| Mapk3 | 128 | Casp3 | 21 | Ep300 | 8 | Nos3 | 7 |
| Il6 | 128 | Tyrobp | 19 | Cox8a | 6 | Ins1 | 4 |
| Cdc2 | 113 | Apoe | 18 | Jun | 6 | Cd4 | 4 |
| Vegfa | 105 | Stat3 | 17 | Met | 5 | Dusp1 | 4 |
| Pcna | 101 | Csf1r | 16 | Atf3 | 5 | Egr1 | 3 |
| Cd4 | 99 | Lyn | 16 | Mdm4 | 5 | Dvl3 | 3 |
| Calm1 | 96 | Gfap | 16 | Ccnl1 | 4 | Nfatc2 | 3 |
| Fn1 | 95 | Sqle | 15 | Cyc1 | 4 | Pofut2 | 2 |
| Fos | 91 | Cox7a2 | 15 | Tpr | 4 | Tfip11 | 2 |
d3, day 3; wk1, week 1; wk2, week 2; m1, month 1.
TFs
TFs among the DEGs at the four time-points were identified (Table IV). The greatest number of TFs was identified at d3, including ATF3, JUN and EGR1. The lowest number of TFs was identified at m1, including EGR1 and JUN. Combined with the PPI network, ATF3, EGR1 and JUN were the most important TFs associated with the development of SCI.
Table IV.
Transcription factors among DEGs at four time-points following spinal cord injury in rats.
| Time-points | Transcription factors
|
|
|---|---|---|
| Upregulated gene(s) | Downregulated gene(s) | |
| d3 | Atf3, Crem, Jun, Maf, Nfe2l2, Pax6, Rest, Tceb3, Tfec, Ybx1 | Atf4, Bmyc, Dbp, Egr1, Mef2d, Nr1d1, Nr3c2, Nr4a2, Olig1, Pou6f1, Rxrg, Thrb, |
| wk1 | Rest | Nr3c2, Olig1 |
| wk2 | Atf3, Csrp3, Fosl1, Jun | |
| m1 | Egr1, Jun | |
DEG, differentially expressed gene; d3, day 3; wk1, week 1; wk2, week 2; m1, month 1.
Discussion
At present, no completely restorative treatments for SCI are available (3,30). The present study was performed in order to explore potential biomarkers and molecular mechanisms underlying SCI using bioinformatic methods. Thousands of DEGs were identified by comparing the gene expression profiles of samples from d3, wk1, wk2 and m1 post-SCI with those of healthy Ctrl samples. DEGs were shown to be enriched in pathways associated with immune response, nervous system diseases and cancer. According to the PPI network for d3, wk1, wk2 and m1, a number of hub nodes were identified, including CD4, STAT3 and JUN. TFs were identified in the DEGs, including ATF3, EGR1, OLIG1 and JUN. These genes may be involved in the mechanisms underlying regeneration or self-repair following SCI.
Siebert et al (16) have demonstrated that there is a strong regenerative response during the early stages of SCI. The present study found that OLIG1 was differentially expressed at d3 and wk1 but not at wk2 or m1. OLIG1 is expressed during the maturation and regeneration of human oligodendrocytes (31). Arnett et al (32) demonstrated that OLIG1 is associated with CNS repair in mice. Therefore, OLIG1 may participate in early regenerative responses to SCI. ATF3 is a member of the mammalian activation transcription factor protein family and was found to be differentially expressed at d3, wk1 and wk2. ATF3 was suggested to be a useful marker for regenerative response following nerve root injury (33) and a novel indicator of nerve injury (34). ATF3 is able to bind with other members of the ARF/CREB family, including ATF2, c-JUN and JUNB, and form dimers, which exert transcriptional activation and inhibitory effects (35). In the present study, ATF3 and JUN were present in the PPI network. ATF3 and c-Jun induces the anti-apoptotic factor Hsp27 (36), which activates protein kinase B, thereby inhibiting apoptosis and inducing nerve elongation. The results of the present study suggested that ATF3- and c-JUN-induced Hsp27 expression may be a novel neuron survival response to nerve injury.
A number of immune response-associated DEGs and pathways were identified in the present study. CD4 is a membrane glycoprotein, which is associated with the T-cell receptor signaling pathway. CD4 may be involved in neuronal damage associated with infectious and immune-mediated diseases of the CNS (37,38). STAT3 is activated via phosphorylation in response to various cytokines and growth factors such as interleukins (ILs). STAT3 is associated with a number of chemokine signaling pathways, including the IL-9 signaling pathway, immune response IL-23 signaling pathway and certain pathways associated with cancer. SCI or amyotrophic lateral sclerosis damages spinal motor neurons and forms a glial scar, which prevents neural regeneration. STAT3 is involved in astrogliogenesis and scar formation, and therefore, modulation of STAT3 signaling may be useful for controlling the excessive gliogenic environment and neural repair in patients with SCI (39). In the present study, STAT3 was significantly upregulated at wk1 and wk2, but not at d3, which suggested that there was a regenerative effect associated with STAT3 expression. In addition, RAC2, which exhibited the highest degree of expression at d3, regulates a number of cellular responses and is associated with neutrophil immunodeficiency syndrome (40). Immune responses maintain neurogenesis in adult germinal centers of the damaged CNS, even under non-pathological conditions (41). Treatments to decrease inflammatory responses are likely to be beneficial to CNS recovery following SCI.
The DEGs identified in the present study at four time-points following SCI are involved in a number of disease-associated pathways, including those associated with certain cancers. These results are in accordance with those of other studies. For example, Myers et al (13) demonstrated that patients with SCI exhibited higher morbidity of the cardiovascular system with greater incidences of diabetes, compared with healthy patients. van den Berg et al (42) found that cancer and bacterial infection may enhance SCI. Furthermore, SCI may induce cardiovascular disease and alter immune responses. In addition, immune-associated pathways were predominantly observed during the early stages of SCI (d3, wk1). By contrast, pathways associated with cancer were predominantly observed in the later stage of SCI (wk2). The results of the present study suggested that genes associated with myocardial contraction and immune response may be involved in the mechanisms underlying early-stage SCI.
In conclusion, a number of SCI regeneration-associated genes have been identified using a computational bioinformatics analysis of gene expression, including OLIG1, ATF3 and JUN. The involvement of inflammation in SCI was investigated and associated genes were highlighted, including CD4, STAT3 and RAC2. Furthermore, the results of the present study suggested that SCI may be associated with a number of diseases, including cardiovascular disease and cancers. The present study provided novel insight into the molecular mechanisms of SCI regeneration, which may aid in the development of strategies to enhance recovery following SCI. Further investigations using a larger sample size should be performed to confirm the results of the present study. Since the present study was based on microarray data alone, further studies should incorporate different data types.
References
- 1.National Spinal Cord Injury Statistical Center Spinal cord injury facts and figures at a glance. J Spinal Cord Med. 2013;36:1–2. doi: 10.1179/1079026813Z.000000000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu J. China spinal cord injury network: changes from within. Lancet Neurol. 2009;8:606–607. doi: 10.1016/S1474-4422(09)70162-0. [DOI] [PubMed] [Google Scholar]
- 3.Thuret S, Moon LD, Gage FH. Therapeutic interventions after spinal cord injury. Nat Rev Neurosci. 2006;7:628–643. doi: 10.1038/nrn1955. [DOI] [PubMed] [Google Scholar]
- 4.Freund P, Weiskopf N, Ashburner J, et al. MRI investigation of the sensorimotor cortex and the corticospinal tract after acute spinal cord injury: a prospective longitudinal study. Lancet Neurol. 2013;12:873–881. doi: 10.1016/S1474-4422(13)70146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald JW, Sadowsky C. Spinal-cord injury. Lancet. 2002;359:417–425. doi: 10.1016/S0140-6736(02)07603-1. [DOI] [PubMed] [Google Scholar]
- 6.Mariano ED, Batista CM, Barbosa BJ, et al. Current perspectives in stem cell therapy for spinal cord repair in humans: a review of work from the past 10 years. Arq Neuropsiquiatr. 2014;72:451–456. doi: 10.1590/0004-282X20140051. [DOI] [PubMed] [Google Scholar]
- 7.Young W. Spinal cord regeneration. Cell Transplant. 2014;23:573–611. doi: 10.3727/096368914X678427. [DOI] [PubMed] [Google Scholar]
- 8.Liu NK, Xu XM. Phospholipase A2 and its molecular mechanism after spinal cord injury. Mol Neurobiol. 2010;41:197–205. doi: 10.1007/s12035-010-8101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly DJ, Popovich PG. Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp Neurol. 2008;209:378–388. doi: 10.1016/j.expneurol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Na W, Lee JY, et al. Molecular mechanism of Jmjd3-mediated interleukin-6 gene regulation in endothelial cells underlying spinal cord injury. J Neurochem. 2012;122:272–282. doi: 10.1111/j.1471-4159.2012.07786.x. [DOI] [PubMed] [Google Scholar]
- 11.Kerschensteiner M, Gallmeier E, Behrens L, et al. Activated human T cells, B cells and monocytes produce brain-derived neurotrophic factor in vitro and in inflammatory brain lesions: a neuroprotective role of inflammation? J Exp Med. 1999;189:865–870. doi: 10.1084/jem.189.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yekutiel M, Brooks M, Ohry A, Yarom J, Carel R. The prevalence of hypertension, ischaemic heart disease and diabetes in traumatic spinal cord injured patients and amputees. Paraplegia. 1989;27:58–62. doi: 10.1038/sc.1989.9. [DOI] [PubMed] [Google Scholar]
- 13.Myers J, Lee M, Kiratli J. Cardiovascular disease in spinal cord injury: an overview of prevalence, risk, evaluation and management. Am J Phys Med Rehabil. 2007;86:142–152. doi: 10.1097/PHM.0b013e31802f0247. [DOI] [PubMed] [Google Scholar]
- 14.Groah SL, Weitzenkamp DA, Lammertse DP, Whiteneck GG, Lezotte DC, Hamman RF. Excess risk of bladder cancer in spinal cord injury: evidence for an association between indwelling catheter use and bladder cancer. Arch Phys Med Rehabil. 2002;83:346–351. doi: 10.1053/apmr.2002.29653. [DOI] [PubMed] [Google Scholar]
- 15.Kalisvaart JF, Katsumi HK, Ronningen LD, Hovey R. Bladder cancer in spinal cord injury patients. Spinal Cord. 2010;48:257–261. doi: 10.1038/sc.2009.118. [DOI] [PubMed] [Google Scholar]
- 16.Siebert JR, Middelton FA, Stelzner DJ. Intrinsic response of thoracic propriospinal neurons to axotomy. BMC Neurosci. 2010;11:69. doi: 10.1186/1471-2202-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai J, He X, Wang F, et al. Gene expression signature analysis and protein-protein interaction network construction of spinal cord injury. Eur Rev Med Pharmacol Sci. 2013;17:2941–2948. [PubMed] [Google Scholar]
- 18.Jin L, Wu Z, Xu W, et al. Identifying gene expression profile of spinal cord injury in rat by bioinformatics strategy. Mol Biol Rep. 2014;41:3169–3177. doi: 10.1007/s11033-014-3176-8. [DOI] [PubMed] [Google Scholar]
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [DOI] [Google Scholar]
- 23.Warnes GR, Bolker B, Bonebakker L, et al. gplots: Various R programming tools for plotting data. R package version 2.7.4. 2009. [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database Issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtine G, van den Brand R, Musienko P. Spinal cord injury: Time to move. Lancet. 2011;377:1896–1898. doi: 10.1016/S0140-6736(11)60711-3. [DOI] [PubMed] [Google Scholar]
- 31.Othman A, Frim DM, Polak P, Vujicic S, Arnason BG, Boullerne AI. Olig1 is expressed in human oligodendrocytes during maturation and regeneration. Glia. 2011;59:914–926. doi: 10.1002/glia.21163. [DOI] [PubMed] [Google Scholar]
- 32.Arnett HA, Fancy SP, Alberta JA, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- 33.Lindå H, Sköld MK, Ochsmann T. Activating transcription factor 3, a useful marker for regenerative response after nerve root injury. Front Neurol. 2011;2:30. doi: 10.3389/fneur.2011.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flatters S. ATF3: novel signpost for nerve injury. Neuroreport. 2000;11:A7. doi: 10.1097/00001756-200010200-00003. [DOI] [Google Scholar]
- 35.Koh IU, Lim JH, Joe MK, et al. AdipoR2 is transcriptionally regulated by ER stress-inducible ATF3 in HepG2 human hepatocyte cells. Febs J. 2010;277:2304–2317. doi: 10.1111/j.1742-4658.2010.07646.x. [DOI] [PubMed] [Google Scholar]
- 36.Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci. 2003;23:5187–5196. doi: 10.1523/JNEUROSCI.23-12-05187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohm AP, Carpentier PA, Anger HA, Miller SD. Cutting edge: CD4+ CD25+ regulatory T cells suppress antigen-specific autoreactive immune responses and central nervous system inflammation during active experimental autoimmune encephalomyelitis. J Immunol. 2002;169:4712–4716. doi: 10.4049/jimmunol.169.9.4712. [DOI] [PubMed] [Google Scholar]
- 38.Liblau RS, Gonzalez-Dunia D, Wiendl H, Zipp F. Neurons as targets for T cells in the nervous system. Trends Neurosci. 2013;36:315–324. doi: 10.1016/j.tins.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Natarajan R, Singal V, Benes R, et al. STAT3 modulation to enhance motor neuron differentiation in human neural stem cells. PLoS One. 2014;9:e100405. doi: 10.1371/journal.pone.0100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ambruso DR, Knall C, Abell AN, et al. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci USA. 2000;97:4654–4659. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziv Y, Avidan H, Pluchino S, Martino G, Schwartz M. Synergy between immune cells and adult neural stem/progenitor cells promotes functional recovery from spinal cord injury. Proc Natl Acad Sci. 2006;103:13174–13179. doi: 10.1073/pnas.0603747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Berg ME, Castellote JM, de Pedro-Cuesta J, Mahillo-Fernandez I. Survival after spinal cord injury: A systematic review. J Neurotrauma. 2010;27:1517–1528. doi: 10.1089/neu.2009.1138. [DOI] [PubMed] [Google Scholar]