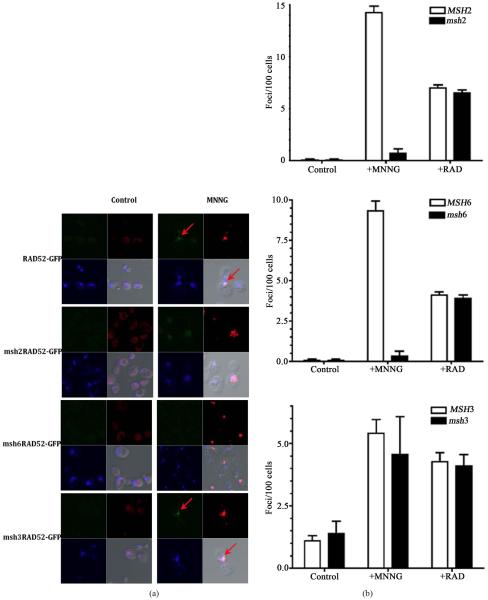
Figure 3.
The MSH2-MSH6 complex is required for foci formation upon DNA damage. (a) Visualization of DNA damage sites as indicated by histone H2AX phosphorylation (red regions). MNNG treated or untreated (control) cells were fixed immediately after treatment with MNNG and processed for direct immunofluorescence microscopy using yeast γ-H2AX antibody as described in the Materials and Methods. Foci of Rad52-GFP (green spots) are indicated by an arrow. Nuclei were demarked by DAPI-stained (blue); (b) The numbers of RAD52-GFP foci formed was determined from 3 independent experiments visualizing at least 400 live yeast cells per experiment. Error bars indicate standard deviation. Strains used were wild type (RAD52-GFP), msh2 mutants (msh2 RAD52-GFP), msh3 mutants (msh3 RAD52-GFP) and msh6 mutants (msh6RAD52-GFP).