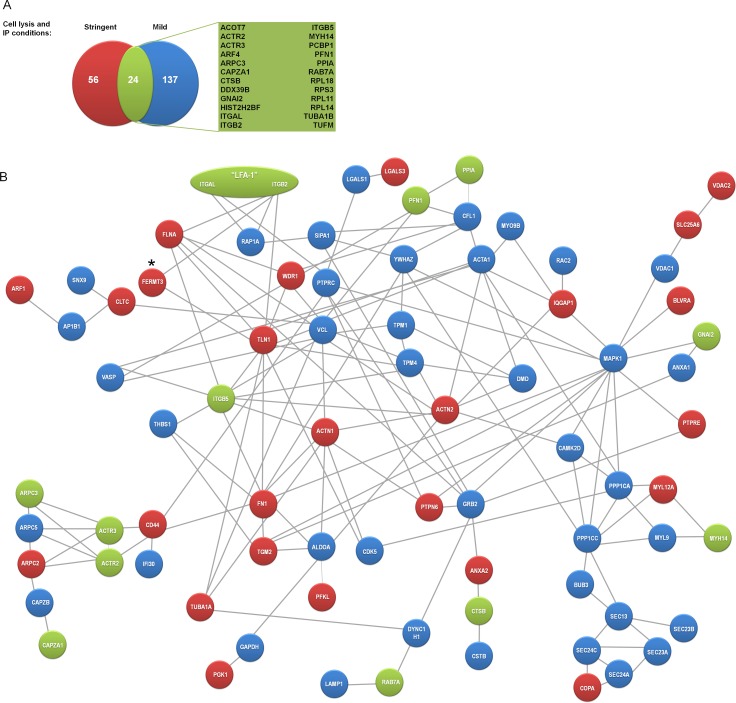
Fig 5. Protein-protein interaction network of directly interacting LFA-1 binding candidates derived from DCs.
(A) Comparison of immunoprecipitated LFA-1 from day 6 imDCs in mild and stringent lysis conditions. Venn diagrams of proteins identified in DCs in stringent (red) and mild (blue) IP conditions. Numbers of identified proteins, as well as common proteins (yellow) are indicated. (B) A network of LFA-1 (heterodimer formed by an αL (ITGAL) and β2 (ITGB2) chain) binding partners was generated by fusing the data sets derived from mild and stringent IP conditions in DCs, uploading the protein names to the database of functional protein interactions (STRING v9.05) and retrieving experimental proven direct protein-protein interactions. (ribosomal and histone complexes were removed for better visualization of proteins involved in integrin function). The resulting network was redrawn by the authors. Based on our MS data, we could construct a high confidence network (score 0.6) containing 78 nodes and 154 connections. Blue nodes represent proteins identified in mild lysis conditions, and red nodes represent proteins identified in stringent lysis conditions. Green nodes represent proteins identified both in mild and stringent IP condition. * indicates an interaction that was not present in the STRING database (version 9.1) with experimental support, but this node and interactions were added by the authors based on the current literature [27].