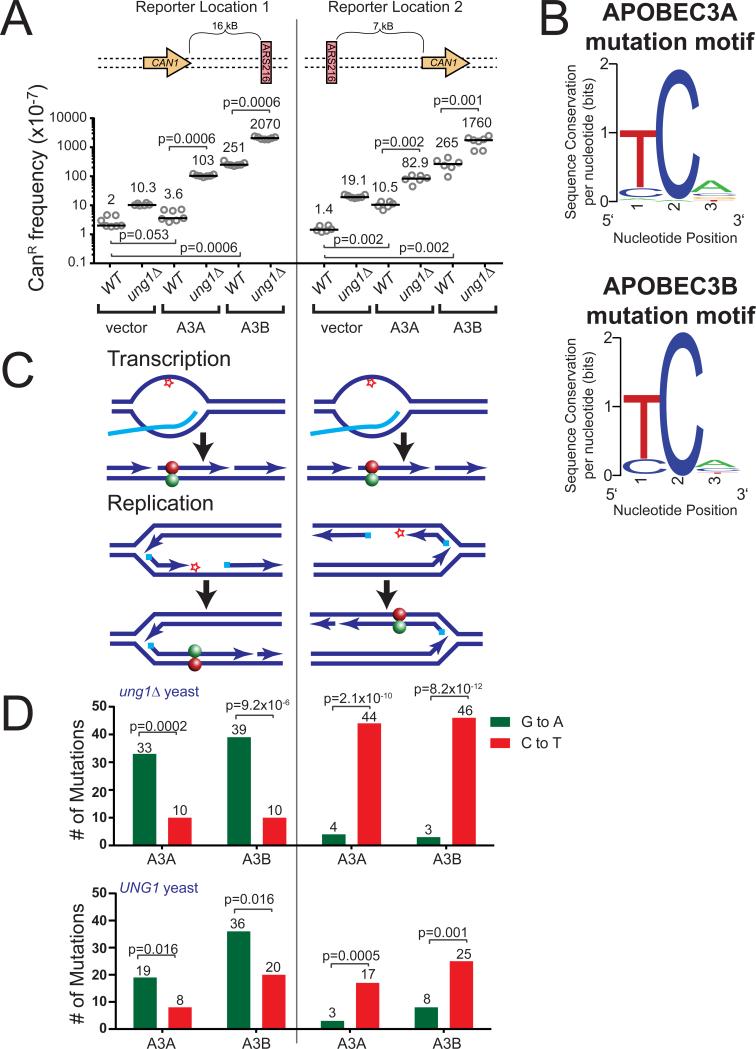
Figure 1. APOBEC3A and 3B induce strand-bias mutations around ARS216.
(A) Frequencies of canavanine-resistance (CanR) induced in WT and ung1Δ yeast following transformation with vector control plasmid, A3A expression plasmid, or A3B expression plasmid. CanR frequency was determined in isogenic strains with the CAN1 gene located either 16 kB 5′ of the origin of replication, ARS216, or 7 kB 3′ of ARS216. Horizontal bars and numeric values indicate the median frequency of six or seven independent replicates. Statistical significance was determined by two-tailed non-parametric Mann-Whitney Rank test. See also Figure S1. (B) Sequence logo describing the favored trinucleotide sequences mutated by A3A (top) and A3B (bottom) in ung1Δ yeast. See also Table S1. (C) Specific deamination of ssDNA formed in a transcription bubble would result in C to T transitions (red ball) within CAN1 regardless of whether the gene is positioned 5′ or 3′ of ARS216. In contrast, deamination of ssDNA formed during lagging strand synthesis would result in G to A substitutions (green ball) 5′ and C to T substitutions 3′ or the origin. (D) The number of G to A (green) and C to T (red) substitutions induced in the CAN1 gene (located 5′ and 3′ of ARS216) by A3A and A3B in ung1Δ and UNG1 yeast as determined by sequencing independent CanR isolates. P-values were determined using a one-tailed g-test goodness-of-fit comparing the ratio of C to T substitutions to G to A substitutions to an expected 1:1 ratio. See also Table S1.