Abstract
MicroRNAs (miRs) have been causally implicated in the progression and development of a wide variety of cancers. miRs modulate the activity of key cell signaling networks by regulating the translation of pathway component proteins. Thus, the pharmacological targeting of miRs that regulate cancer cell signaling networks, either by promoting (using miR-supplementation) or by suppressing (using anti-sense oligonucleotide based strategies) miR activity is an area of intense research. The RAS-Extracellular signal regulated kinase (ERK) pathway represents a major miR-regulated signaling network that endows cells with some of the classical hallmarks of cancer, and is often inappropriately activated in malignancies by somatic genetic alteration through point mutation or alteration of gene copy number. In addition, recent progress indicates that many tumors may be deficient in GTPase activating proteins (GAPs) due to the collaborative action of oncogenic microRNAs. Recent studies also suggest that in tumors harboring a mutant RAS allele there is a critical role for wild type RAS proteins in determining overall RAS-ERK pathway activity. Together, these two advances comprise a new opportunity for therapeutic intervention. In this review, we evaluate miR-based therapeutic strategies for modulating RAS-ERK signaling in cancers, in particular for more direct modulation of RAS-GTP levels, with the potential to complement current strategies in order to yield more durable treatment responses. To this end, we discuss the potential for miR-based therapies focused on three prominent miRs including the pan-RAS regulator let-7 and the GAP regulator comprised of miR-206 and miR-21 (miR-206/21).
Keywords: microRNAs, RAS-ERK signaling, RAS-GTP, cancer
INTRODUCTION
MicroRNAs (miRs) are genomically encoded single stranded noncoding RNAs that are typically 19–25 nucleotides (nt) in length and result from extensive processing of endogenous hairpin-shaped precursors [He and Hannon, 2004; Kim, 2005; Chen and Rajewsky, 2007; Ameres and Zamore, 2013]. miRs were initially identified in C. elegans as gene products required for the regulation of proper developmental timing [Wightman et al., 1991; Lee et al., 1993]. Subsequently, thousands of miRs and putative miR-encoding genes have been identified in a wide variety of organisms, including in plants and metazoans. Since their discovery, miRs have emerged as critical regulators of gene expression and cell signaling, and are functionally implicated in numerous cellular processes including development, differentiation, proliferation, and apoptosis [Kasinski and Slack, 2011; Sayed and Abdellatif, 2011; Iorio and Croce, 2012; Sun and Lai, 2013]. As a consequence of these critical roles, dysregulation of miR expression, activity, and signaling results in a multitude of pathological states, including the development and progression of cancers [Esquela-Kerscher and Slack, 2006; Calin and Croce, 2006a; Calin and Croce, 2006b].
Perturbation of key signaling networks endows cells with many of the well-established hallmarks of cancer, such as enhanced cell proliferation, resistance to cell stress and death, and motility, and is implicated in the pathogenesis of virtually every type of human malignancy [Hanahan and Weinberg, 2000; Weinberg, 2007]. Attributed to elevated levels of membrane-associated RAS-GTP, signaling downstream of RAS proto-oncogenes through the RAF-MEK-ERK mitogen activated protein kinase (MAPK) pathway is often inappropriately activated in a wide variety of cancers, promoting several of the classical hallmarks of cancer [Bos, 1989; Schubbert et al., 2007; Bos et al., 2007; Karnoub and Weinberg, 2008; Young et al., 2009; Tidyman and Rauen, 2009; Pylayeva-Gupta et al., 2011]. Activation of this hierarchically tiered signaling pathway can occur through a variety of ways, including in response to stimulation by upstream inputs (i.e., receptor tyrosine kinases (RTKs), integrins, ion channels, etc.), somatic mutation of pathway components such as RAS and RAF, and alteration of the expression of pathway regulators [Schlessinger, 2000; Johnson and Lapadat, 2002; Downward, 2003; Kolch, 2005; Dhillon et al., 2007; Mebratu and Tesfaigzi, 2009]. In cancer cells, the activation of RAS-ERK signaling has been most prominently documented in the context of somatic acquisition of activating point mutations in RAS GTPase genes (e.g., KRAS, HRAS, NRAS). These mutations render the encoded gene products resistant to the inhibitory action of GTPase activating proteins (GAPs, which potently stimulate GTP hydrolysis by RAS) [Trahey and Mccormick, 1987; Boguski and Mccormick, 1993; Scheffzek et al., 1995; Scheffzek et al., 1997; Bos et al., 2007]. In addition to GAPs, numerous factors contribute to the proper spatio-temporal regulation of RAS-ERK signaling, including guanine nucleotide exchange factors (GEFs), which promote recycling to the active, GTP bound state by reducing the affinity of RAS proteins for GDP. In addition, other proteins function as scaffolds or adaptors for the proper localization of signaling molecules, such as SPRED1 which is critical for the membrane localization of NF1/GAP [Bos et al., 2007; McKay and Morrison, 2007; Wortzel and Seger, 2011; Stowe et al., 2012; Roskoski, Jr., 2012a; Roskoski, Jr., 2012b]. More recently, analyses of tumor cells containing a RAS mutation indicated that the wild type proteins encoded by the remaining, unmutated RAS alleles play a critical role in pathway output, identifying these wild type proteins as a potential Achilles’ heel for therapeutic targeting [Jeng et al., 2012; Young et al., 2013; Grabocka et al., 2014; Sharma et al., 2014]
miRs represent yet another level of regulatory control of RAS-ERK signaling and, in certain tumor cells such as basal-like or triple-negative breast cancer (TNBC), can represent major regulators of RAS-ERK activity by impacting the translation of pathway components such as GAPs and/or GAP-associated scaffolding proteins such as SPRED1 [Johnson et al., 2005; Fish et al., 2008; Thum et al., 2008; Hatley et al., 2010; Sun et al., 2013; Sharma et al., 2014; Stark et al., 2015b]. Unraveling how miRs impact RAS-ERK signaling in cancer has the potential to uncover novel therapeutic strategies which can complement conventional modalities and/or targeted therapies such as kinase inhibitors. In this review, we briefly describe miR biogenesis and how miRs can impact the pathogenesis of cancer by altering cell signaling. We discuss miR-based therapeutic strategies and necessary considerations for the successful use of in vivo miR-targeting agents. We then describe the structure of the circuitry of the RAS-ERK signaling pathway, and briefly review the utility of inhibiting this pathway in the treatment of cancers. We consider how miRs can regulate RAS-ERK signaling by targeting specific pathway components and critical regulatory proteins, including wild-type RAS proteins. We next evaluate the prospect of targeting miR-mediated regulation of RAS-ERK in the therapy of cancers and contrast this therapeutic modality with other pharmacological RAS-ERK inhibitory strategies. We conclude by proposing a therapeutic strategy for the more direct suppression of RAS-GTP levels, including the in vivo silencing of the cooperative GAP regulators, miR-206 and miR-21 (i.e., miR-206/21). Recent studies indicate that these two miRs maintain RAS-ERK signaling in breast cancer cells by limiting the translation of a major GAP termed RASA1 and the NF1/GAP-associated factor, SPRED1 [Sharma et al., 2014]. In TNBC cells the resulting suppression of GAP activities by endogenous miR-206/21 is critical for the maintenance of wild type RAS-GTP levels, RAS-ERK signaling and malignant properties not only for cancer cells harboring wild type RAS proteins, but similarly in tumor cells harboring a RAS mutation, further supporting a critical role of wild type RAS in cells harboring the mutant protein [Jeng et al., 2012; Young et al., 2013; Grabocka et al., 2014; Sharma et al., 2014].
MICRORNAS
miR Biogenesis
In humans, the predominant miR biosynthetic route involves the transcription of miR-encoding genes by RNA polymerase II [He and Hannon, 2004; Kim, 2005; Chen and Rajewsky, 2007; Ameres and Zamore, 2013]. The resulting primary miR transcripts (pri-miRs) are processed in the nucleus by the RNAse-III enzyme Drosha, which exists as part of a heterodimeric complex with DGCR8 microprocessor complex unit (also known as “DiGeorge Syndrome Critical Region 8”) to yield stem-loop hairpin structures of approximately 70 nt in length, termed precursor miRs (pre-miRs) [Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004; Landthaler et al., 2004; Han et al., 2004]. Pre-miRs are exported out of the nucleus predominantly by the action of Ran-GTPase/exportin-5 and further processed by the cytoplasmic RNAse-III enzyme Dicer, to yield RNA duplexes composed of the mature miRs of approximately 19–25 nt in length [Bernstein et al., 2001; Ketting et al., 2001; Provost et al., 2002; Lee et al., 2002; Lee et al., 2003]. These are termed miR-miR* duplexes. Following strand selection, “miR” represents the more abundant strand of the duplex and the less abundant strand is denoted “miR*” [Ambros et al., 2003; Griffiths-Jones et al., 2006]. In addition to this major route of biogenesis, some miRs arise from alternate synthetic pathways, including processes that do not utilize RNA polymerase II mediated transcription, or the action of Drosha/DGCR8 or Dicer proteins [Yang and Lai, 2011]. For example, miR-genes located near Alu-repeat sequences or tRNA genes can be transcribed by RNA polymerase III [Borchert et al., 2006]. Furthermore, miRs can also arise from a Drosha/DGCR8 independent synthetic process from the splicing of intronic regions from mRNA transcripts and are termed miRtrons [Okamura et al., 2007]. Finally, miR-miR* duplexes may be produced by direct cleavage of RNA precursors by the endoribonuclease Argonaute-2 (AGO2) to yield mature miRs in a Dicer independent process [Yang et al., 2010].
miR-mediated Regulation of Protein Translation
Mature miRs typically repress the translation of mRNA transcripts by associating with the catalytic center (AGO endoribonucleases) of multiprotein complexes termed RNA-induced silencing complexes (RISCs) [Bartel, 2009; Czech and Hannon, 2011]. miR-miR* duplexes are subsequently unwound and a mature miR strand is retained in RISC based on the relative thermodynamic stability across the miR-miR* duplex. The miR* strand may subsequently either be cleaved or be ejected from the assembled complex. The mature RISC then scans target mRNA sequences. The selectivity for target mRNAs arises from miR sequence complementarity to portions of the target mRNAs, particularly in the 3′ untranslated region (3′ UTR). In mammals, sequence complementarity between bases 2–8 near the 5′ end of the miR (termed the miR-seed sequence) and portions of the mRNA is the dominant factor that guides RISCs to target and to repress the translation of specific transcripts [He and Hannon, 2004; Chen and Rajewsky, 2007; Jackson et al., 2010; Ameres and Zamore, 2013]. The resulting miR-mediated translational repression may occur by (i) blocking translation initiation, (ii) enhanced mRNA degradation, or by (iii) site specific cleavage of the target mRNA, though the latter process occurs infrequently in mammals [He and Hannon, 2004; Chen and Rajewsky, 2007; Jackson et al., 2010; Ameres and Zamore, 2013]. Interestingly, though miRs typically repress the translation of target mRNAs, a few miRs have been documented to promote the translation of cognate transcripts, suggesting an additional level of complexity in this modality of regulating gene expression [Vasudevan and Steitz, 2007; Vasudevan et al., 2007; Vasudevan et al., 2008; Steitz and Vasudevan, 2009; Jangra et al., 2010; Lin et al., 2011; Ameres and Zamore, 2013].
miRs are capable of widespread regulation of gene expression, with more than 60% of protein coding mRNA transcripts possessing at least one evolutionarily conserved miR seed complementary sequence [Ha and Kim, 2014]. Conversely, whereas the translation of a single mRNA transcript may be impacted by multiple miRs, individual miRs can potentially regulate the expression of hundreds of protein coding transcripts, each of which may have diverse cellular functions. Importantly, the collaborative action of miRs can potently modulate the activity of key signaling networks by targeting one or more pathway components [He and Hannon, 2004; Chen and Rajewsky, 2007; Jackson et al., 2010; Ameres and Zamore, 2013]. miRs may also participate in reciprocal regulation of transcripts that are critical for miR biogenesis or maturation, and thus constitute components of feedback loops along with their target mRNAs.
These complex miR-mRNA interaction paradigms are essential for the maintenance of homeostasis of critical cell physiological processes and the perturbation of miR regulation of important cell signaling networks, such as the RAS-ERK pathway, contributes to a wide variety of pathological states, including cancer. Indeed, recent work suggests that miR signaling to RAS-ERK can be responsible for maintaining low GAP activity that leads to high RAS-ERK pathway activity, regardless of the RAS mutational status [Sharma et al., 2014]. Furthermore, studies indicate that mutant RAS-GTP level does not on its own determine pathway activity, but instead that both the mutant and wild type RAS proteins must preferentially associate with GTP in order to maintain higher levels of pathway activity [Jeng et al., 2012; Young et al., 2013; Grabocka et al., 2014; Sharma et al., 2014].
miR-based Therapeutic Strategies
As miR dysregulation is a prominent feature of many pathological states, the therapeutic targeting of specific miRs holds the promise for therapy of various diseases [Kasinski and Slack, 2010; Inui et al., 2010; Kasinski and Slack, 2011; Stenvang et al., 2012; Ling et al., 2013; Li and Rana, 2014; van and Kauppinen, 2014]. miR-based therapeutics are defined as strategies that restore or inhibit miR function to counteract perturbations in miR-signaling. These strategies include (i) restoring miR function by supplementation of miR-mimics, (ii) inhibiting miR function by synthetic anti-sense oligonucleotide-based approaches targeting endogenous miRs (termed anti-miRs and antagomiRs) or by (iii) modulating miR function by non-oligonucleotide based methods including peptide nucleic acids (PNAs) [Kasinski and Slack, 2011; Ling et al., 2013; Li and Rana, 2014]. Furthermore, oligonucleotide based strategies may feature various chemical modifications to enhance the stability and affinity of these therapeutic agents [Kasinski and Slack, 2011; Ling et al., 2013; Li and Rana, 2014].
Despite our extensive knowledge of attractive miR targets in various disease states, the successful utilization of miR-based therapeutic strategies in vivo is challenged by several obstacles [Kasinski and Slack, 2011; Ling et al., 2013; Li and Rana, 2014]. First, the successful delivery of these agents to the target tissue is limited by physical, anatomical, pharmacokinetic, and pharmacodynamic barriers, which may be overcome in part by a variety of delivery methods and targeting strategies including miR-cholesterol conjugation, liposome encapsulation, miR-nanoparticle conjugation, and antibody- or aptamer-based targeting methods [Kasinski and Slack, 2011; Ling et al., 2013; Li and Rana, 2014]. As miR-mimics and anti-miRs can be degraded by ubiquitously present nucleases in the human body, synthetic modification of these agents is necessary for their stability [Lamond and Sproat, 1993; Cummins et al., 1995].
Second, miR-based therapies, including anti-sense strategies, have the potential for promiscuous miR inhibition, yielding off-target effects [Kasinski and Slack, 2011; Ling et al., 2013; Li and Rana, 2014]. miR-therapeutics must be able to discern between the many miRs that may share identical/similar seed sequences with the intended target. Thus, careful consideration of target miRs must be undertaken to ensure that miR-based therapeutics can modulate the intended endogenous target with a high degree of specificity, to minimize off-target effects.
Finally, the administration of miR-therapeutics and carrier vehicles such as targeted nanoparticles may cause deleterious consequences by miR sequence independent off-target effects [Kasinski and Slack, 2011; Ling et al., 2013; Li and Rana, 2014]. miR agents may be detected by both the innate and adaptive arms of the human immune systems, and chemical modification to therapeutic components is necessary to avoid immunostimulatory off-target effects [Judge et al., 2005; Hornung et al., 2005]. Furthermore, these agents may alter physiological processes (e.g., blood coagulation, complement cascade activation) or may induce organ system dysfunction (e.g., hepatotoxicity, nephrotoxicity), owing to the accumulation, clearance, and excretion of these molecules [Galbraith et al., 1994; Henry et al., 1997; Swayze et al., 2007]. Therefore, the consideration of these therapeutic barriers is necessary for the successful utilization of miR-based therapeutics.
RAS-ERK SIGNALING
Organization and Regulation of the RAS-ERK Signaling Pathway
RAS-ERK signaling is a critical mediator of cell physiological processes including cell proliferation, differentiation and motility [Johnson and Lapadat, 2002; Downward, 2003; Kolch, 2005; Dhillon et al., 2007; Young et al., 2009; Mebratu and Tesfaigzi, 2009; Pylayeva-Gupta et al., 2011]. Activation of this pathway occurs downstream of signaling inputs including receptor tyrosine kinases (RTKs), integrins, and ion channels, which are in turn activated by a variety of stimuli and cell stresses [Schlessinger, 2000; Johnson and Lapadat, 2002; Lemmon and Schlessinger, 2010]. The RAS family of GTPases consists of four members (HRAS, NRAS, KRAS4A and KRAS4B [alternatively spliced variants]) that arise from three distinct genes (HRAS, NRAS, and KRAS) and occupy a critical position in relaying signaling from these diverse inputs to activate downstream effector pathways such as the RAF-MEK-ERK pathway, as well as the phosphoinositide 3-kinase (PI3-K)-AKT pathway [McCubrey et al., 2006; Roberts and Der, 2007; Young et al., 2009]. RAS proteins fulfill this important role by functioning as binary switches that alternate between the GTP-bound “on” state (RAS-GTP), which enables RAS to engage downstream effector pathways, and the GDP-bound “off” state (RAS-GDP) [Bos et al., 2007; Vigil et al., 2010]. The activation state of RAS is predominantly governed by critical accessory proteins that enable the transition between either of these states. Essential factors for proper RAS signaling include guanine nucleotide exchange factors (GEFs) which promote the formation of RAS-GTP, GAPs, which promote GTP hydrolysis, and scaffolding proteins such as SPRED1 which appears critical for membrane localization of NF1/GAP [Stowe et al., 2012].
In humans, the activation of the RAF-MEK-ERK pathway is initiated by the preferential interaction of membrane-associated RAS-GTP with the RAS-binding domain of the RAF family of serine/threonine kinases [Karnoub and Weinberg, 2008; Cox and Der, 2010; Pylayeva-Gupta et al., 2011]. Membrane-associated RAS proteins exist as dimers, and this dimerization may be critical for the activation of RAF kinases (composed of three paralogs: ARAF, BRAF, and CRAF/RAF-1), which occurs in a complex multi-step process [Inouye et al., 2000; Guldenhaupt et al., 2012; Lin et al., 2014]. Activated RAF kinases phosphorylate and activate the dual specificity kinases MEK 1 and MEK 2 (MEK 1/2), which in turn phosphorylate and activate ERK 1 and ERK 2 (ERK 1/2), the terminal effector kinases of this pathway [Johnson and Lapadat, 2002; Roskoski, Jr., 2012a; Roskoski, Jr., 2012b]. In contrast to the limited substrate specificity of RAF and MEK 1/2, ERK 1/2 are capable of phosphorylating and consequently modulating the activity of a wide variety of cytoplasmic and nuclear substrates. Importantly, the activity of ERK 1/2 responsive transcription factors is critical in orchestrating cell responses to numerous input stimuli that lie upstream of RAS GTPases [Roskoski, Jr., 2012a; Roskoski, Jr., 2012b].
Regulation of RAS-ERK Pathway Activity
The activation of RAS-ERK signaling is tightly regulated through a variety of means. RAS-ERK pathway activity is maintained by a delicate balance between factors that promote pathway activation (i.e., GEFs), factors that inhibit pathway activation (i.e., GAPs, DUSPs), and proteins that function as scaffolds, adaptors, and/or provide docking sites for pathway regulatory components [Boguski and Mccormick, 1993; Bos et al., 2007; McKay and Morrison, 2007; Yoshimura, 2009; Cox and Der, 2010; Vigil et al., 2010; Wortzel and Seger, 2011; Roskoski, Jr., 2012a; Roskoski, Jr., 2012b]. These factors may confer signaling specificity to membrane subdomains, allowing distinct effects of the different RAS family members. Additionally, ERK 1/2 can directly phosphorylate and inhibit the activity of the GEF SOS1, CRAF, and MEK1, and thus attenuate signaling by feedback inhibition [Ueki et al., 1994; Rossomando et al., 1994; Buday et al., 1995]. Furthermore, ERK 1/2 can also regulate the transcription of upstream drivers of RAS-ERK signaling such as RTKs [Sears et al., 2000; Amit et al., 2007; Lemmon and Schlessinger, 2010; Duncan et al., 2012].
Tightly controlled spatio-temporal regulation of RAS-ERK signaling is critical for the proper execution of cell physiological processes, and inappropriate regulation of the pathway results in a variety of disease states, including developmental disorders and cancer [Bos et al., 2007; McKay and Morrison, 2007; Wortzel and Seger, 2011; Roskoski, Jr., 2012a; Roskoski, Jr., 2012b]. For example, somatic activating point mutation of RAS and BRAF genes occur in approximately 15–30% and 7–8% of all cancers respectively [Davies et al., 2002; Downward, 2003; Schubbert et al., 2007; Karnoub and Weinberg, 2008; Young et al., 2009; Tidyman and Rauen, 2009; Cox and Der, 2010; Pylayeva-Gupta et al., 2011; Fernandez-Medarde and Santos, 2011]. Though some malignancies, such as pancreatic ductal adenocarcinomas, colorectal carcinomas, and melanomas feature a high proportion of activating RAS and RAF mutations [Bos, 1989; Pylayeva-Gupta et al., 2011], other cancers such as TNBCs display RAS-ERK pathway activation despite the infrequent occurrence of somatic point mutations, thus implicating dysregulation of RAS-ERK through other means [Mirzoeva et al., 2009; Hoeflich et al., 2009b; Cancer Genome, 2012]. Interestingly, cancers such as TNBCs frequently display genetic alterations such as gene copy number changes in pathway components and altered expression of pathway regulatory proteins [van Beers et al., 2005; Herschkowitz et al., 2007; Rakha et al., 2008; Hu et al., 2009; Cancer Genome, 2012; Balko et al., 2012; Balko et al., 2013]. Remarkably, 32% of basal-like breast cancers display KRAS gene amplifications and 30% of cancers of this subtype harbor BRAF gene amplifications [Cancer Genome, 2012].
The Emerging Paradigm of a Critical Role for Wild Type RAS Proteins in Cells Harboring RAS Mutations
Early seminal studies identified the potent transforming ability of virally encoded RAS genes and subsequently characterized these gene products as mutated versions of the human RAS homologs [Harvey, 1964; Kirsten and Mayer, 1967; Scolnick et al., 1973; Scolnick and Parks, 1974; Shih et al., 1979; Krontiris and Cooper, 1981; Santos et al., 1982; Chang et al., 1982; Parada et al., 1982; Der et al., 1982]. Furthermore, the observation of similarly mutated endogenous RAS genes in human tumor samples was critical for our understanding of the molecular basis of carcinogenesis [Capon et al., 1983; Feig et al., 1984; Santos et al., 1984]. These important early studies uncovered that RAS mutations predominantly occur in codons 12, 13, and 61 and that these mutant proteins were constitutively bound to GTP [Taparowsky et al., 1982; Reddy et al., 1982; Tabin et al., 1982; Hall et al., 1983]. Furthermore, mutant RAS possessed far less intrinsic GTPase activity compared to the non-mutant counterparts and were virtually resistant to the action of GAPs [Sweet et al., 1984; Gibbs et al., 1984; McGrath et al., 1984; Manne et al., 1985; Trahey and Mccormick, 1987; Mccormick et al., 1991; Mccormick, 1992]. The striking effect of activated RAS in these early experiments and the identification of RAS mutations in cancers sparked several decades of research that has vastly broadened our knowledge of cell signaling and its role in neoplasia.
But compelling questions regarding the regulation of RAS signaling still remain. Recently, how the activity of wild-type RAS proteins contributes to downstream pathway activation in the context of RAS-mutant cells was uncovered in a series of genetic and biochemical studies [Jeng et al., 2012; Young et al., 2013; Grabocka et al., 2014], including a study by our lab focused on the role of miR-regulated GAPs in RAS-mutant cells [Sharma et al., 2014]. These studies uncovered a previously unappreciated role for wild-type RAS proteins, as well as GEFs and GAPs, as critical signaling molecules in the context of mutant-RAS associated phenotypes. Possibly attributed to the formation of wild type/mutant RAS dimers, these studies found that GEFs and GAPs are critical regulators of tumorigenesis of RAS-mutant cells through their modulation of WT-RAS-GTP levels. These studies concluded that regardless of the RAS mutational status, the ultimate signaling output is likely determined by the ratio of RAS-GTP to RAS-GDP, where the pool of WT-RAS plays a major role, even in cells with mutant RAS. A critical difference between wild-type and mutant RAS proteins is the greater dependence of WT-RAS on GEFs and GAPs [Bos et al., 2007; Vigil et al., 2010]. Consequently these new studies identify these regulators as critical therapeutic targets regardless of the RAS mutational status. These studies establish a new paradigm of how RAS signaling is regulated and highlight the potential of small molecule modulators of GAP or GEF activity.
Therapeutic Targeting of RAS-ERK Signaling in Cancers
Given the important role of RAS-ERK signaling in the development and progression of many cancers, the successful therapeutic inhibition of this pathway has been a long standing goal of the targeted chemotherapy era [Downward, 2003; Karnoub and Weinberg, 2008; Gysin et al., 2011; Pylayeva-Gupta et al., 2011; Mattingly, 2013]. Numerous strategies to inhibit RAS-ERK signaling have been envisioned, including those that (i) directly target RAS, (ii) modulate factors that regulate RAS activity, and those that (iii) target downstream kinases (e.g. RAF, MEK, and ERK). One of the most promising therapeutic strategies, utilizing ATP analogues as allosteric or competitive inhibitors of RAF or MEK kinase activity has proceeded toward clinical utility, but with prolonged therapeutic responses limited by a variety of factors [Liu et al., 2013; Johnson et al., 2014]. Despite the potent action of these compounds in vitro, therapeutic resistance emerges rapidly and hampers the successful use of these kinase inhibitors [Engelman et al., 2007; Garrett et al., 2011; Chandarlapaty et al., 2011; Duncan et al., 2012; Johnson et al., 2014; Stuhlmiller et al., 2015].
Contributing to this resistance, acute loss of RAS-ERK signaling in cancer cells results in adaptive reprogramming, including reprogramming of the kinome, with upregulation of multiple (receptor tyrosine) kinases (RTKs) including PDGFRβ, DDR1, and others [Engelman et al., 2007; Garrett et al., 2011; Chandarlapaty et al., 2011; Duncan et al., 2012; Johnson et al., 2014; Stuhlmiller et al., 2015]. In addition, pathway inhibition by agents such as MEK inhibitor is thwarted by the loss of negative feedback regulation, including ERK 1/2 mediated inhibition of positive pathway regulators such as MEK1 and BRAF [Duncan et al., 2012; Johnson et al., 2014]. Another factor is that phosphorylation of MEK1 by cRAF results in reduced affinity of MEK 1/2 allosteric inhibitors [Ueki et al., 1994; Ritt et al., 2010]. Furthermore, and particularly in melanoma, long term treatment with RAS-ERK inhibitory compounds results in tumor cells acquiring somatic mutations in NRAS, MEK2, or AKT1 to counteract sustained inhibited signaling [Nazarian et al., 2010; Villanueva et al., 2013; Shi et al., 2014]. Finally, through the induction of multiple RTKs, the activation of alternative signaling pathways (e.g., PI3-K-AKT) can compensate for the inhibited RAS-ERK signaling [Sos et al., 2009; Hoeflich et al., 2009a; Nazarian et al., 2010; Duncan et al., 2012; Sun et al., 2014]. Either singly or in combination, these adaptive changes ultimately circumvent blocked signaling and prevent sustained therapeutic responses. Thus, the development and optimization of more effective RAS-ERK pathway inhibitory strategies and counteracting the rapid emergence of resistant signaling represent critical obstacles to effective therapeutic intervention.
A major goal has been more direct methods for suppression of RAS-GTP levels [Gysin et al., 2011; Mattingly, 2013]. Toward this end, the recent recognition that WT RAS proteins play a critical role in pathway output would appear to reenergize ongoing efforts to target GEFs and GAPs [Bos et al., 2007; Vigil et al., 2010]. Other ongoing approaches include reovirus-based therapies and siRNA therapy against mutant KRAS [Thirukkumaran and Morris, 2009; Zorde et al., 2013; Yuan et al., 2014; Golan et al., 2015].
MICRORNA REGULATION OF MUTANT AND WILD TYPE RAS-GTP
The past decade has provided considerable insight into the critical regulatory roles that miRs exert over key cancer relevant signaling networks such as the RAS-ERK pathway. Our knowledge of how miRs can modulate RAS-ERK pathway activation continues to grow as potential miR-mRNA regulatory networks are identified using a variety of strategies including in silico miR target prediction methods, profiling of the cellular transcriptome/proteome, and experimental validation of putative interactors that typically employs translational reporter assays.
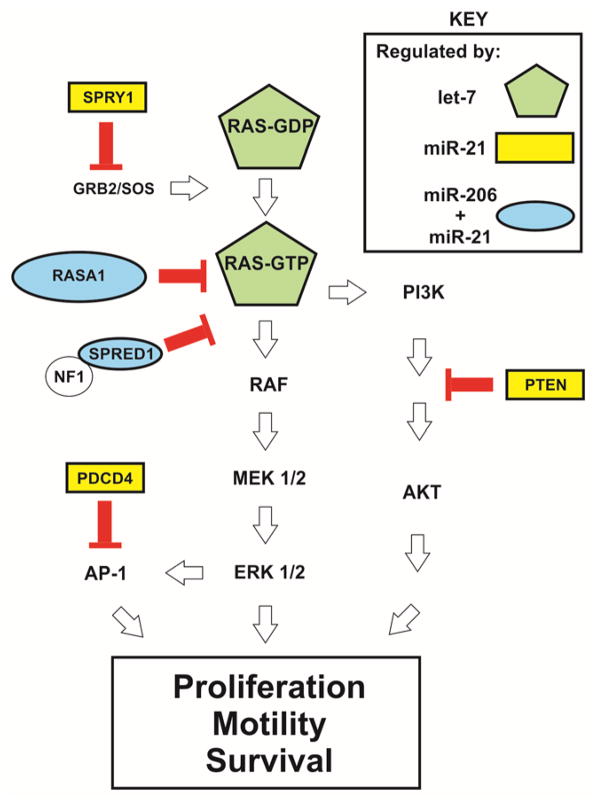
Three major paradigms of miR-mediated RAS-ERK regulation have emerged from these studies. miRs can impact the translation of (i) core RAS-ERK pathway components (e.g., let-7 targets HRAS, NRAS, and KRAS) [Johnson et al., 2005], (ii) critical pathway regulatory proteins that are required for the proper spatio-temporal control of RAS-ERK signaling (e.g., miR-206 and/or miR-21 collaboratively target RASA1, SPRED1, SPRY1; and miR-21 individually targets PTEN) (Fig. 1) [Meng et al., 2007; Hatley et al., 2010; Sharma et al., 2014], and (iii) upstream drivers and downstream effector/regulatory molecules (e.g., miR-9-3p targets ITGB1, and miR-206/21 co-target PDCD4) [Zawistowski et al., 2013; Lin et al., 2015]. Examples of miRs that regulate RAS-ERK pathway activity in a variety of cancer contexts are listed in Table 1. Indeed, these miRs represent potential therapeutic substrates and targets that can be modulated in the treatment of cancer. In the following sections, we evaluate the therapeutic potential of three miRs, let-7 and miR-206/21, that hold great promise as potential therapeutic targets in the treatment of cancers by impacting RAS-ERK signaling.
Figure 1. miRs regulate RAS-ERK pathway activity by regulation of RAS-GTP.
The schematic shows the organization of the RAS-ERK pathway. miR-206/21 co-targeted repressors of RAS-ERK signaling are indicated in ovals. The GAP protein NF1 is indicated as a likely catalytic partner of SPRED1 [Stowe et al., 2012]. The let-7 miR targets each of the RAS family GTPases including KRAS, HRAS, and NRAS.
Table 1.
miRs that regulate RAS-ERK pathway activity in a variety of cancer contexts
| miR | Target(s) | Disease contexts | References |
|---|---|---|---|
| let-7 | HRAS, NRAS, KRAS, cMYC | Multiple cancer contexts including lung adenocarcinoma | [Johnson et al., 2005] [Johnson et al., 2007] [Yu et al., 2007] [Kasinski and Slack, 2010] [He et al., 2010] |
| miR-21 | RASA1, SPRED1, SPRY1, SPRY2, PTEN, PDCD4 | Multiple cancer contexts including TNBCs | [Iorio et al., 2005] [Volinia et al., 2006] [Frankel et al., 2008] [Jin et al., 2013] [Mei et al., 2013] [Wickramasinghe et al., 2009] [Asangani et al., 2008] [Meng et al., 2006] [Sharma et al., 2014] |
| miR-206 | RASA1, SPRED1, PDCD4 | TNBCs | [Sharma et al., 2014] [Lin et al., 2015] |
| miR-31 | RASA1 | Colorectal carcinoma | [Sun et al., 2013] |
| miR-143 | NRAS | Glioma | [Wang et al., 2014] |
| miR-514a | NF1 | Melanoma | [Stark et al., 2015a] |
| miR-223 | RASA1 | Colorectal adenocarcinoma | [Sun et al., 2015] |
| miR-181a | KRAS | Oral squamous cell carcinoma | [Shin et al., 2011] |
| miR-524-5p | BRAF, ERK2 | Melanoma | [Liu et al., 2014] |
| miR-96 | KRAS | Pancreatic adenocarcinoma | [Yu et al., 2010] |
| miR-30c | KRAS | Breast cancer | [Tanic et al., 2012] |
Let-7 Represses the Translation of the RAS Family of GTPases
The let-7 gene was initially identified as an essential regulator of patterning development in the nematode C. elegans, and was among the first defined miRs [Wightman et al., 1991; Lee et al., 1993]. Subsequent studies observed evolutionary conservation of let-7 and identified related paralogs in the genomes of multiple species, including humans [Pasquinelli et al., 2000; Reinhart et al., 2000]. Similarly to C. elegans, human let-7 is critical for epithelial cellular differentiation and proliferation [Yu et al., 2007; Johnson et al., 2007]. Furthermore, reduced expression of let-7 in cancer occurs through genetic deletion, mutation, epigenetic silencing, and post-transcriptional regulation of let-7 biogenesis, and decreased let-7 expression has been implicated in pathogenesis of a wide variety of malignancies, including cancers of the lung, colon, ovary, and breast [Johnson et al., 2005; Yu et al., 2007; Kasinski and Slack, 2010]. These studies suggest a tumor suppressive function for this miR.
Prominent mechanisms by which let-7 exerts a tumor suppressive role is by repressing the translation of the three RAS proteins (HRAS, NRAS, and KRAS) and cMYC, a downstream effector of RAS-ERK signaling [Johnson et al., 2005; He et al., 2010]. Studies analyzing in vitro and in vivo models of non-small cell lung cancer (NSCLC), as well as human tumor samples, show that let-7 expression is inversely correlated with the expression of KRAS, a critical promoter of NSCLC tumorigenesis. Let-7 abrogates tumor development and RAS-ERK signaling in an autochthonous model of NSCLC driven by activated KRAS (KRASG12D) [Esquela-Kerscher et al., 2008; Kumar et al., 2008]. Consistent with this previous result, a tumor suppressive role for let-7 was observed in a study analyzing a xenograft model of NSCLC [He et al., 2010]. Additionally, in a breast cancer context, let-7 antagonizes the maintenance, survival, and self-renewal of cancer stem-like cells (CSCs), and this suppressive activity was correlated with the reduced expression of RAS and HMGA2 [Yu et al., 2007]. Thus, by suppressing RAS expression, let-7 can attenuate RAF-MEK-ERK signaling and dependent oncogenic phenotypes regardless of the RAS-mutation status of cancers. These studies suggest that let-7 can act as both a cancer-preventative and cancer-therapeutic agent, and point to let-7 supplementation as a promising strategy to target RAS-ERK signaling in the treatment of cancers.
MiR-206/21 Collaboratively Repress the Translation of RASA1 and SPRED1 and Inhibit GAP Activity
Our laboratory recently identified two miRs, miR-206 and the well characterized oncogene miR-21 (collectively: miR-206/21), as critical regulators of RAS-ERK signaling in TNBC cells (Fig. 1) [Sharma et al., 2014]. Whereas miR-206 is well characterized in regulating the differentiation of adult muscle stem cells, the role of endogenous miR-206 in breast cancer is less well known [Kim et al., 2006; Chen et al., 2010; Dey et al., 2011; Liu et al., 2012]. In contrast to miR-206, miR-21 is prominently upregulated in many malignancies, including in breast cancer, and promotes tumorigenesis by repressing the translation of multiple tumor suppressors, including negative regulators of RAS-ERK signaling (e.g., SPRY1, RASA1, and PDCD4), as well as RAS-PI3K signaling (e.g., PTEN) [Iorio et al., 2005; Volinia et al., 2006; Meng et al., 2006; Frankel et al., 2008; Thum et al., 2008; Asangani et al., 2008; Wickramasinghe et al., 2009; Jin et al., 2013; Mei et al., 2013].
We found that the expression of miR-206/21 was dependent on the zinc-finger pluripotency factor Kruppel-like factor 4 (KLF4), which has most often been implicated as a poor prognostic factor in breast cancer [Pandya et al., 2004; Kamalakaran et al., 2011; Chen et al., 2012]. Whereas KLF4 and miR-206 are preferentially expressed in MaCSCs, miR-21 is similarly expressed in these two compartments, consistent with the “on/off” mode of miR-21 regulation by KLF4 [Sharma et al., 2014; Lin et al., 2015]. Furthermore, recent studies from our laboratory indicate that KLF4 and/or its dependent miRs are important regulators of anti-apoptotic signaling in breast cancer cells and promote survival against diverse forms of stress, including treatment with conventional cytotoxic or targeted chemotherapies [Farrugia et al., 2015; Lin et al., 2015].
The combined action of miR-206/21 promotes signaling by repressing the translation of multiple RAS-ERK pathway inhibitory proteins which act at various hierarchical levels in this signaling network [Sharma et al., 2014]. Interestingly, the manipulation of each individual miR did not yield large changes in pathway activity, suggesting that the collaborative action of miR-206/21 was required to achieve a substantial effect. Indeed, treatment of TNBC cells with anti-miR-206/21 was sufficient to suppress pathway activity by greater than 80%. We found that miR-206/21 co-target and co-suppress the translation of the GAP RASA1, and the Neurofibromatosis 1 (NF1) GAP associated protein, SPRED1. This GAP-deficient state interferes with RAS inactivation (i.e., the formation of signaling-deficient WT-RAS-GDP), and consequently promotes RAS-ERK signaling, RAS dependent cell phenotypes, and TNBC tumorigenesis. Importantly, whereas inhibition of KLF4-miR-206/21 signaling potently suppresses RAS-ERK signaling in multiple RAS-mutant TNBC models (MDA-MB-231– KRASG13D, Hs578t – HRASG12D, and SUM159PT – HRASG12D) as well as in cells that exclusively harbor WT-RAS proteins, stable shRNA-mediated suppression of RASA1 and SPRED1 promoted pathway activity on its own, and rendered cells virtually resistant to anti-miR-206/21. Consequently, TNBC cells and potentially many other tumor types are GAP-deficient owing to the collaborative action of miR-206/21 on RASA1 and SPRED1. Therefore the RASA1 and SPRED1 transcripts represent latent tumor suppressors with the potential for reactivation by anti-miR-206/21. Our analysis of GAP signaling and RAS-GTP levels in this study yielded results that were consistent with the newly emerging paradigm that the output of RAS-ERK signaling is critically dependent on the activation status of WT-RAS, and uncovered a previously unappreciated role for GAP proteins in cells harboring RAS mutations [Jeng et al., 2012; Young et al., 2013; Grabocka et al., 2014; Sharma et al., 2014].
The Potential for in vivo Inhibition of MiR-206/21 for the Treatment of Cancer
In addition to the numerous studies that have elucidated the oncogenic role of miR-21, our analysis of KLF4-miR signaling suggests that inhibition of miR-206/21 has the potential to be a promising therapeutic strategy in the treatment of TNBC and in other cancers. In vivo silencing of miR-206/21 using anti-sense oligonucleotide based strategies could effectively attenuate RAS-ERK signaling by more directly suppressing RAS-GTP levels. Improved therapeutic effects may be achieved when used in combination with other pathway inhibitory strategies or in conjunction with cytotoxic chemotherapy. Furthermore, other strategies for suppression of oncogenic miRs hold promise, including aptamer-mediated inhibition of nucleolin [Pichiorri et al., 2013], a cell surface protein required for the maturation of a specific subset of miRs, including miRs that promote RAS-ERK signaling (e.g., miR-21, miR-221).
Therapeutic targeting of these miRs in combination with other therapeutic modalities could offer an advantage over single kinase inhibition strategies. Due to the rapid adaptive reprogramming and the emergence of inhibitor-resistant signaling, these do not yield durable responses [Engelman et al., 2007; Garrett et al., 2011; Chandarlapaty et al., 2011; Duncan et al., 2012; Johnson et al., 2014; Stuhlmiller et al., 2015]. Probably owing to the more physiologic enhancement of GAP activity, suppression of miR-206/21 leads to potent inhibition of RAS-ERK signaling and RAS-dependent phenotypes [Sharma et al., 2014], but appears not to destabilize c-MYC or to induce the dramatic adaptive reprogramming response that results from kinase inhibitors (unpublished observations, SBS and JMR). Additionally, miR-206/21 suppression may sensitize cells to the effects of other anti-cancer therapeutic modalities. We have recently found that miR-206 represses the translation of the pro-apoptotic protein PDCD4, a well established miR-21 target, to protect breast cancer cells from apoptosis in response to cytotoxic chemotherapy [Lin et al., 2015]. Thus the combinatorial suppression of miR-206/21 could potentiate the effects of conventional treatments and thereby reduce the required dosage of these agents, potentially mitigating the adverse events associated with anti-cancer therapy.
While therapeutic windows are notoriously difficult to predict from the analysis of models, mice deficient in either miR-206 or miR-21 develop normally and appear healthy as adults, supporting the potential for dual inhibition as a therapeutic strategy. Thus, the therapeutic inhibition of miR-206/21 activity has the potential to target RAS-ERK signaling through the re-expression of GAP activity, and holds great promise for the treatment of RAS-driven cancers such as TNBC.
CONCLUSIONS
The RAS-ERK signaling pathway is critical in the development and progression of numerous malignancies, and more effective targeted pathway inhibition has the potential to greatly improve the treatment of cancers. Despite extensive work that has culminated in the development of numerous pathway inhibitory strategies, direct suppression of RAS-GTP levels has been difficult to achieve, and the successful utilization of existing kinase inhibitory agents is hampered by adaptive reprogramming of cell signaling. The consistent demonstration of the inefficacy of single kinase inhibition strategies has prompted the consideration of alternative routes of targeting RAS-ERK signaling components and pathway regulatory molecules. miRs represent one such set of important regulatory molecules that can be targeted for the therapeutic inhibition of pathway activity. These miR-based therapeutic strategies involve the supplementation of RAS-ERK inhibitory miRs using miR-mimics (e.g., let-7), or the in vivo silencing of miRs that promote pathway activity using anti-sense strategies (e.g., miR-206/21). Unlike single kinase inhibitors, these targeted miR-based therapeutic strategies may yield durable anti-tumor responses as they can collaboratively target multiple levels of this signaling pathway and regulate other cell physiologic processes that are critical to RAS-ERK mediated tumorigenesis. Though development and optimization of improved miR delivery methods is necessary, targeting RAS-ERK signaling by miR-based therapeutics holds great promise in the treatment of cancers that are reliant on this signaling pathway.
Acknowledgments
This work was supported by grants NCI RO1 CA127405 (to JMR), the Jo and Ben Statler Chair in Breast Cancer Research, and the Wilmer V. and Helen B. Morley Memorial Fund at the Mary Babb Randolph Cancer Center (MBRCC).
References
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, Sanders ME, Granja-Ingram NM, Smith JJ, Meszoely IM, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012;18:1052–1059. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, Gomez H, Cook RS, Arteaga CL. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res. 2013;73:6346–6358. doi: 10.1158/0008-5472.CAN-13-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Mccormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bos JL. RAS oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Buday L, Warne PH, Downward J. Downregulation of the Ras activation pathway by MAP kinase phosphorylation of Sos. Oncogene. 1995;11:1327–1331. [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006b;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006a;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cancer Genome AN. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon DJ, Seeburg PH, McGrath JP, Hayflick JS, Edman U, Levinson AD, Goeddel DV. Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature. 1983;304:507–513. doi: 10.1038/304507a0. [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EH, Furth ME, Scolnick EM, Lowy DR. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982;297:479–483. doi: 10.1038/297479a0. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Lin SE, Lin YM, Lin SH, Chen DR, Chen CL. Association of Expression of Kruppel-like Factor 4 and Kruppel-like Factor 5 with the Clinical Manifestations of Breast Cancer. Pathol Oncol Res. 2012;18:161–168. doi: 10.1007/s12253-011-9422-7. [DOI] [PubMed] [Google Scholar]
- Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Farrugia MK, Sharma SB, Lin CC, McLaughlin SL, Vanderbilt DB, Ammer AG, Salkeni MA, Stoilov P, Agazie YM, Creighton CJ, et al. Regulation of anti-apoptotic signaling by Kruppel-like factors 4 and 5 mediates lapatinib resistance in breast cancer. Cell Death Dis. 2015;6:e1699. doi: 10.1038/cddis.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA, Bast RC, Jr, Knapp RC, Cooper GM. Somatic activation of rasK gene in a human ovarian carcinoma. Science. 1984;223:698–701. doi: 10.1126/science.6695178. [DOI] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Galbraith WM, Hobson WC, Giclas PC, Schechter PJ, Agrawal S. Complement activation and hemodynamic changes following intravenous administration of phosphorothioate oligonucleotides in the monkey. Antisense Res Dev. 1994;4:201–206. doi: 10.1089/ard.1994.4.201. [DOI] [PubMed] [Google Scholar]
- Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JB, Sigal IS, Poe M, Scolnick EM. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984;81:5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan T, Khvalevsky EZ, Hubert A, Gabai RM, Hen N, Segal A, Domb A, Harari G, David EB, Raskin S, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015 doi: 10.18632/oncotarget.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014;25:243–256. doi: 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kotting C, Gerwert K. N-Ras forms dimers at POPC membranes. Biophys J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysin S, Salt M, Young A, Mccormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hall A, Marshall CJ, Spurr NK, Weiss RA. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983;303:396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- HARVEY JJ. An unidentified virus which causes the rapid production of tumors in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van RE, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He XY, Chen JX, Zhang Z, Li CL, Peng QL, Peng HM. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer Res Clin Oncol. 2010;136:1023–1028. doi: 10.1007/s00432-009-0747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SP, Novotny W, Leeds J, Auletta C, Kornbrust DJ. Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev. 1997;7:503–510. doi: 10.1089/oli.1.1997.7.503. [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, Januario T, Savage H, Punnoose E, Truong T, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009a;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, Januario T, Savage H, Punnoose E, Truong T, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009b;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de FA, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Hu X, Stern HM, Ge L, O’Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J Biol Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol. 2010;84:6810–6824. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng HH, Taylor LJ, Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat Commun. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin XL, Sun QS, Liu F, Yang HW, Liu M, Liu HX, Xu W, Jiang YY. microRNA 21-mediated suppression of Sprouty1 by Pokemon affects liver cancer cell growth and proliferation. J Cell Biochem. 2013;114:1625–1633. doi: 10.1002/jcb.24504. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Stuhlmiller TJ, Angus SP, Zawistowski JS, Graves LM. Molecular pathways: adaptive kinome reprogramming in response to targeted inhibition of the BRAF-MEK-ERK pathway in cancer. Clin Cancer Res. 2014;20:2516–2522. doi: 10.1158/1078-0432.CCR-13-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A, Riggs M, Banerjee N, Synnestvedt M, Schlichting E, et al. DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol. 2011;5:77–92. doi: 10.1016/j.molonc.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Potential microRNA therapies targeting Ras, NFkappaB and p53 signaling. Curr Opin Mol Ther. 2010;12:147–157. [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kirsten WH, Mayer LA. Morphologic responses to a murine erythroblastosis virus. J Natl Cancer Inst. 1967;39:311–335. [PubMed] [Google Scholar]
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- Krontiris TG, Cooper GM. Transforming activity of human tumor DNAs. Proc Natl Acad Sci U S A. 1981;78:1181–1184. doi: 10.1073/pnas.78.2.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Sproat BS. Antisense oligonucleotides made of 2′-O-alkylRNA: their properties and applications in RNA biochemistry. FEBS Lett. 1993;325:123–127. doi: 10.1016/0014-5793(93)81427-2. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- Lin CC, Liu LZ, Addison JB, Ivanov AV, Ruppert JM. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol Cell Biol. 2011;31:2513–2527. doi: 10.1128/MCB.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Sharma SB, Farrugia MK, McLaughlin SL, Ice RJ, Loskutov YV, Pugacheva EN, Brundage KM, Chen D, Ruppert JM. Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival. Oncogenesis. 2015 doi: 10.1038/oncsis.2015.8. in press; to be published online 06/08/2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Iversen L, Tu HL, Rhodes C, Christensen SM, Iwig JS, Hansen SD, Huang WY, Groves JT. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc Natl Acad Sci U S A. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122:2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS. Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SM, Lu J, Lee HC, Chung FH, Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5:9444–9459. doi: 10.18632/oncotarget.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne V, Bekesi E, Kung HF. Ha-ras proteins exhibit GTPase activity: point mutations that activate Ha-ras gene products result in decreased GTPase activity. Proc Natl Acad Sci U S A. 1985;82:376–380. doi: 10.1073/pnas.82.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly RR. Activated Ras as a Therapeutic Target: Constraints on Directly Targeting Ras Isoforms and Wild-Type versus Mutated Proteins. ISRN Oncol. 2013;2013:536529. doi: 10.1155/2013/536529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccormick F. Coupling of ras p21 signalling and GTP hydrolysis by GTPase activating proteins. Philos Trans R Soc Lond B Biol Sci. 1992;336:43–47. doi: 10.1098/rstb.1992.0042. [DOI] [PubMed] [Google Scholar]
- Mccormick F, Martin GA, Clark R, Bollag G, Polakis P. Regulation of ras p21 by GTPase activating proteins. Cold Spring Harb Symp Quant Biol. 1991;56:237–241. doi: 10.1101/sqb.1991.056.01.029. [DOI] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D’Assoro AB, et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul. 2006;46:249–279. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- McGrath JP, Capon DJ, Goeddel DV, Levinson AD. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984;310:644–649. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle. 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z, Zhao RC. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem. 2013;114:1374–1384. doi: 10.1002/jcb.24479. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, et al. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 2009;69:565–572. doi: 10.1158/0008-5472.CAN-08-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pichiorri F, Palmieri D, De LL, Consiglio J, You J, Rocci A, Talabere T, Piovan C, Lagana A, Cascione L, et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J Exp Med. 2013;210:951–968. doi: 10.1084/jem.20120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982;300:149–152. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol Cell Biol. 2010;30:806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012a;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem Biophys Res Commun. 2012b;417:5–10. doi: 10.1016/j.bbrc.2011.11.145. [DOI] [PubMed] [Google Scholar]
- Rossomando AJ, Dent P, Sturgill TW, Marshak DR. Mitogen-activated protein kinase kinase 1 (MKK1) is negatively regulated by threonine phosphorylation. Mol Cell Biol. 1994;14:1594–1602. doi: 10.1128/mcb.14.3.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della PG, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of of BALB- and Harvey-MSV transforming genes. Nature. 1982;298:343–347. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Klebe C, Fritzwolf K, Kabsch W, Wittinghofer A. Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature. 1995;374:378–381. doi: 10.1038/374378a0. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Scolnick EM, Parks WP. Harvey sarcoma virus: a second murine type C sarcoma virus with rat genetic information. J Virol. 1974;13:1211–1219. doi: 10.1128/jvi.13.6.1211-1219.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick EM, Rands E, Williams D, Parks WP. Studies on the nucleic acid sequences of Kirsten sarcoma virus: a model for formation of a mammalian RNA-containing sarcoma virus. J Virol. 1973;12:458–463. doi: 10.1128/jvi.12.3.458-463.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM, Salkeni MA, Ruppert JM. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol. 2014;34:4143–4164. doi: 10.1128/MCB.00480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hong A, Kong X, Koya RC, Song C, Moriceau G, Hugo W, Yu CC, Ng C, Chodon T, et al. A novel AKT1 mutant amplifies an adaptive melanoma response to BRAF inhibition. Cancer Discov. 2014;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C, Shilo BZ, Goldfarb MP, Dannenberg A, Weinberg RA. Passage of phenotypes of chemically transformed cells via transfection of DNA and chromatin. Proc Natl Acad Sci U S A. 1979;76:5714–5718. doi: 10.1073/pnas.76.11.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404:896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, Heynck S, Stuckrath I, Weiss J, Fischer F, et al. Identifying genotype-dependent efficacy of single and combined. Proc Natl Acad Sci U S A. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MS, Bonazzi VF, Boyle GM, Palmer JM, Symmons J, Lanagan CM, Schmidt CW, Herington AC, Ballotti R, Pollock PM, et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget. 2015a doi: 10.18632/oncotarget.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MS, Bonazzi VF, Boyle GM, Palmer JM, Symmons J, Lanagan CM, Schmidt CW, Herington AC, Ballotti R, Pollock PM, et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget. 2015b doi: 10.18632/oncotarget.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA, Vasudevan S. miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans. 2009;37:931–935. doi: 10.1042/BST0370931. [DOI] [PubMed] [Google Scholar]
- Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe IB, Mercado EL, Stowe TR, Bell EL, Oses-Prieto JA, Hernandez H, Burlingame AL, Mccormick F. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, Angus SP, Collins KA, Granger DA, Reuther RA, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep. 2015;11:390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- Sun D, Wang C, Long S, Ma Y, Guo Y, Huang Z, Chen X, Zhang C, Chen J, Zhang J. C/EBP-beta-activated microRNA-223 promotes tumour growth through targeting RASA1 in human colorectal cancer. Br J Cancer. 2015;112:1491–1500. doi: 10.1038/bjc.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1) J Biol Chem. 2013;288:9508–9518. doi: 10.1074/jbc.M112.367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP, Bennett CF. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet RW, Yokoyama S, Kamata T, Feramisco JR, Rosenberg M, Gross M. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature. 1984;311:273–275. doi: 10.1038/311273a0. [DOI] [PubMed] [Google Scholar]
- Tabin CJ, Bradley SM, Bargmann CI, Weinberg RA, Papageorge AG, Scolnick EM, Dhar R, Lowy DR, Chang EH. Mechanism of activation of a human oncogene. Nature. 1982;300:143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- Tanic M, Yanowsky K, Rodriguez-Antona C, Andres R, Marquez-Rodas I, Osorio A, Benitez J, Martinez-Delgado B. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS One. 2012;7:e38847. doi: 10.1371/journal.pone.0038847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky E, Suard Y, Fasano O, Shimizu K, Goldfarb M, Wigler M. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–765. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- Thirukkumaran C, Morris DG. Oncolytic viral therapy using reovirus. Methods Mol Biol. 2009;542:607–634. doi: 10.1007/978-1-59745-561-9_31. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M, Mccormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Ueki K, Matsuda S, Tobe K, Gotoh Y, Tamemoto H, Yachi M, Akanuma Y, Yazaki Y, Nishida E, Kadowaki T. Feedback regulation of mitogen-activated protein kinase kinase kinase activity of c-Raf-1 by insulin and phorbol ester stimulation. J Biol Chem. 1994;269:15756–15761. [PubMed] [Google Scholar]
- van Beers EH, van WT, Wessels LF, Li Y, Oldenburg RA, Devilee P, Cornelisse CJ, Verhoef S, Hogervorst FB, van’t Veer LJ, et al. Comparative genomic hybridization profiles in human BRCA1 and BRCA2 breast tumors highlight differential sets of genomic aberrations. Cancer Res. 2005;65:822–827. [PubMed] [Google Scholar]
- van RE, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, Li B, Swoboda RK, Wilson M, Vultur A, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–1099. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shi ZM, Jiang CF, Liu X, Chen QD, Qian X, Li DM, Ge X, Wang XF, Liu LZ, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014;5:5416–5427. doi: 10.18632/oncotarget.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. Growth Factors, Receptors, and Cancer. In: Weinberg RA, editor. The Biology of Cancer. Garland Science, Taylor and Francis Group, LLC; New York: 2007. pp. 119–158. [Google Scholar]
- Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- Wortzel I, Seger R. The ERK Cascade: Distinct Functions within Various Subcellular Organelles. Genes Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A. Regulation of cytokine signaling by the SOCS and Spred family proteins. Keio J Med. 2009;58:73–83. doi: 10.2302/kjm.58.73. [DOI] [PubMed] [Google Scholar]
- Young A, Lou D, Mccormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- Young A, Lyons J, Miller AL, Phan VT, Alarcon IR, Mccormick F. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, Hsu DJ, Grace D, Carver JO, Zuber J, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 2014;4:1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawistowski JS, Nakamura K, Parker JS, Granger DA, Golitz BT, Johnson GL. MicroRNA 9-3p targets beta1 integrin to sensitize claudin-low breast cancer cells to MEK inhibition. Mol Cell Biol. 2013;33:2260–2274. doi: 10.1128/MCB.00269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorde KE, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]