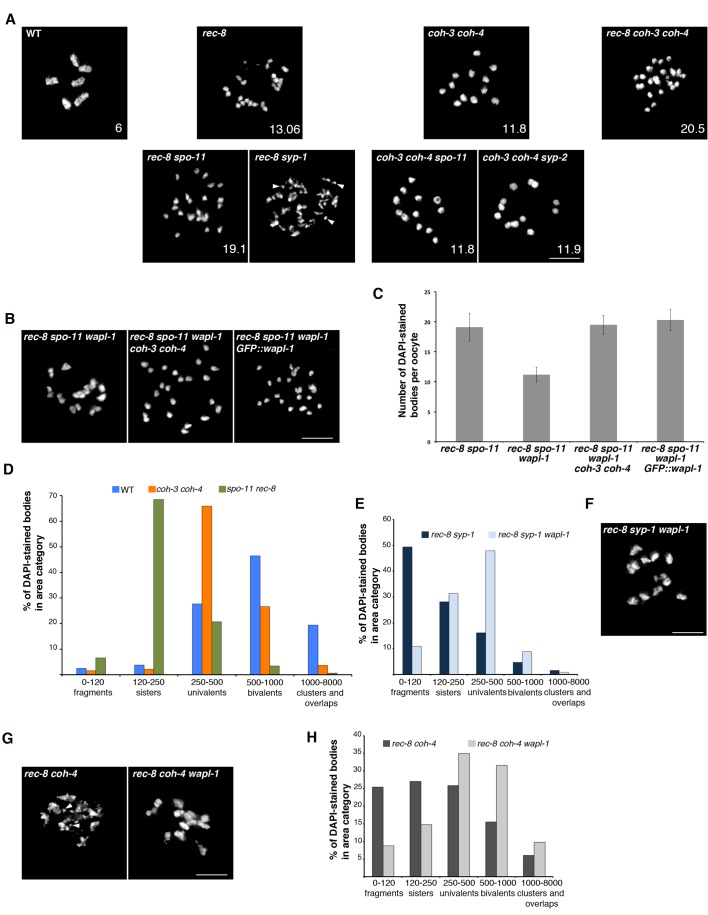
Figure 6. WAPL-1 antagonizes COH-3/4 cohesion in diakinesis oocytes.
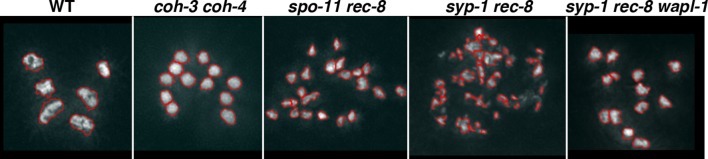
(A) Projections of diakinesis oocytes of the indicated genotypes stained with DAPI. 6 DAPI-stained bodies (WT) indicates presence of 6 bivalents, 12 DAPI-stained bodies indicates absence of chiasmata, and the presence of more than 12 DAPI-stained bodies indicates separation of sister chromatids, with 24 demonstrating separation of all sisters. The average number of DAPI-stained bodies observed in each genotype is indicated on the bottom right of each panel, number of nuclei analyzed: WT (20), rec-8 (29), coh-3 coh-4 (16), rec-8 coh-3 coh-4 (17), rec-8 spo-11 (94), coh-3 coh-4 spo-11 (26), and coh-3 coh-4 syp-2 (26). Arrowheads in rec-8 syp-1 panel point to chromosome fragments. Note that removal of SPO-11 or SYP-1/2 causes loss of cohesion in rec-8 mutants but not in coh-3 coh-4 double mutants. (B) Projections of diakinesis oocytes of the indicated genotypes stained with DAPI, quantification shown in (C). Note the reduction of DAPI-stained bodies in rec-8 spo-11 wapl-1 compared with three other genotypes (p<0.0001 in all cases, t-test). Error bars = standard deviation. Number of nuclei analyzed: rec-8 spo-11 (94), rec-8 spo-11 wapl-1 (85), rec-8 spo-11 wapl-1 coh-3 coh-4 (29), rec-8 spo-11 wapl-1GFP::wapl-1 (27). (D) Automated quantification (CellProfiler) of area sizes corresponding to chromatin bodies in projections of diakinesis oocytes of indicated genotypes stained with DAPI. Values on the X axis represent area in pixels and binning of the different categories was adjusted using oocytes of known phenotypes: WT (bivalents), coh-3 coh-4 (univalents) spo-11 rec-8 (detached sisters). Number of oocytes analyzed: WT (40), coh-3 coh-4 (61) spo-11 rec-8 (116). (E) Automated quantification of area sizes corresponding to chromatin bodies in projections of diakinesis oocytes, note that removing WAPL-1 from rec-8 syp-1 double mutants causes a large decrease of chromosome fragments and an increase in univalents. Number of nuclei analyzed: rec-8 syp-1 (52), rec-8 syp-1 wapl-1 (36). (F) Example of DAPI-stained oocyte from rec-8 syp-1 wapl-1 demonstrating absence of chromosome fragments, compare with rec-8 syp-1 example shown in A. (G) Projections of diakinesis oocytes stained with DAPI, note that chromosome fragments are present in rec-8 coh-4 (arrowheads) but not in rec-8 coh-4 wapl-1 oocytes. (H) Automated quantification of area sizes corresponding to chromatin bodies in projections of diakinesis oocytes, note that removing WAPL-1 from rec-8 coh-4 double mutants causes a large decrease of chromosome fragments. 26 diakinesis oocytes were analyzed for both rec-8 coh-4 and rec-8 coh-4 wapl-1.