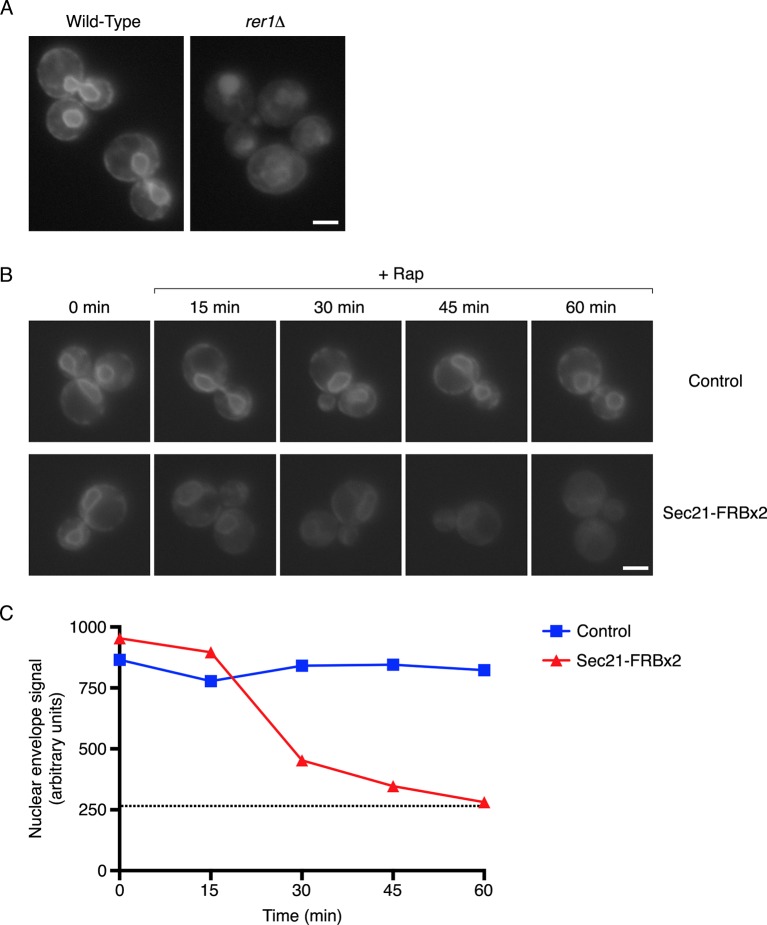
Figure 4. Depletion of Sec71TMD-GFP from the ER after COPI inactivation.
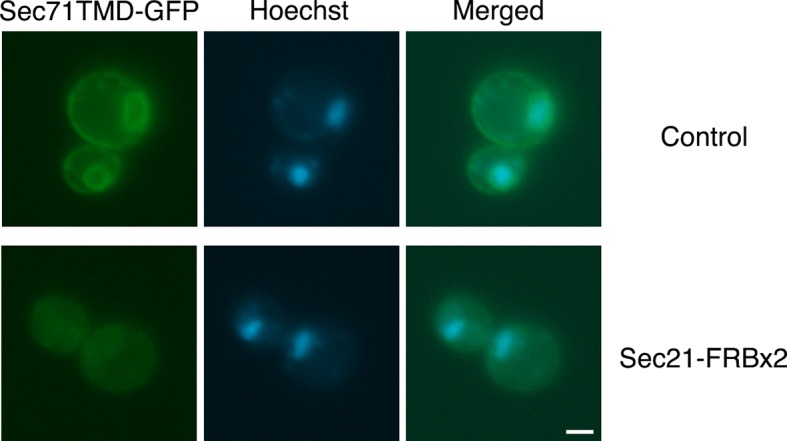
(A) Sec71TMD-GFP requires Rer1 for normal ER localization. An expression vector for Sec71TMD-GFP was integrated into the wild-type JK9-3da strain or into an isogenic rer1△ strain. Cells grown to log phase in NSD at 30°C were compressed beneath a coverslip and visualized in a single focal plane by widefield microscopy. Representative images taken at the same exposure are shown. Scale bar, 2 μm. (B) Anchoring of COPI causes loss of Sec71TMD-GFP from the ER. Both of the strains shown here contained integrating vectors for expressing the mitochondrial anchor OM45-FKBPx4 as well as Sec71TMD-GFP, and one strain also expressed Sec21-FRBx2 to anchor COPI. Cells growing at 30°C were incubated for 10 min with 0.0002% Hoechst 33342 to stain nuclei, followed by an additional treatment for up to 60 min with 1 μg/mL rapamycin (“Rap”). At the indicated time points after rapamycin addition, cells were compressed beneath a coverslip and visualized in a single focal plane by widefield microscopy. Representative images of GFP fluorescence are shown. Scale bar, 2 μm. (C) The data from (B) were quantified by measuring the GFP fluorescence signals from nuclei as a proxy for the ER signals. For this purpose, the signal in the blue channel was used to select nuclei in ImageJ using the Magic Wand tool. These selections were expanded by 12 pixels to include the nuclear envelope, and the GFP fluorescence from the selected areas in the green channel was quantified. Plotted are average fluorescence signals per unit area. Each data point was derived from approximately 40–80 cells, and was adjusted by subtracting an average background signal from nuclei in the parental strain lacking GFP. The dashed line represents the average background-corrected nuclear fluorescence signal from rer1Δ cells that exhibited low to moderate vacuolar fluorescence.
Figure 4—figure supplement 1. Labeling of nuclei to quantify Sec71TMD-GFP fluorescence in the nuclear envelope.