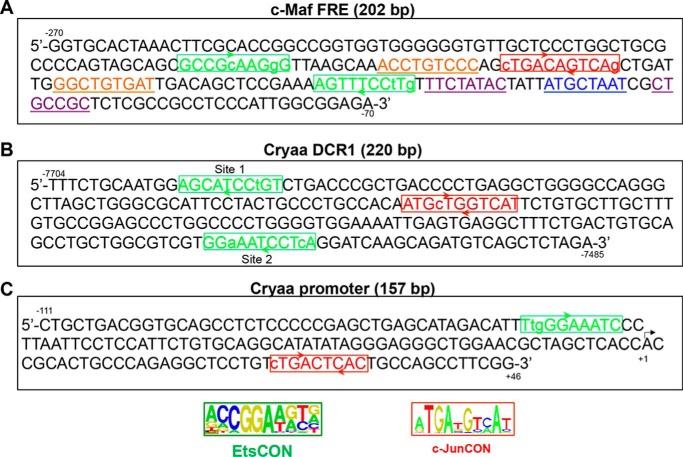
FIGURE 3.
Multiple AP-1- and Ets-binding sites are present in regulatory regions of mouse c-Maf and αA-crystallin genes. A, predicted Ets and AP-1 binding sites within the FRE (−272/−70) of the c-Maf promoter. B, Ets and AP-1 binding sites within the 220 bp DCR1 enhancer of the αA-crystallin locus. C, Ets and AP-1 binding sites within the αA-crystallin promoter (−111/+46). Note that the Ets-binding site matches to a cis-acting region of the mouse Cryaa promoter established earlier (61) and binding of c-Jun to the AP-1 site was reported elsewhere (62). The Ets binding sites were searched by allowing up to two mismatches in the Ets consensus sequence CCGGA(A/T)(A/G)(C/T) (33). AP-1 (c-Jun) binding sites were found by allowing up to two mismatches in the c-Jun palindromic consensus sequence ATGA(T/C)GTCAT (34) or TGA(G/C)T(A/C)A (35). Candidate Smad- (underlined/purple)-sites in the c-Maf FRE were predicted by using Smad consensus motifs 5′-GTCTAGAC-3′(58) and 5′-CWGSMGCY-3′ (57). The candidate Pou2f1 (underlined/blue)-site was predicted by using a consensus motif from the JASPAR database (motif ID MA0785.1) (63, 64).