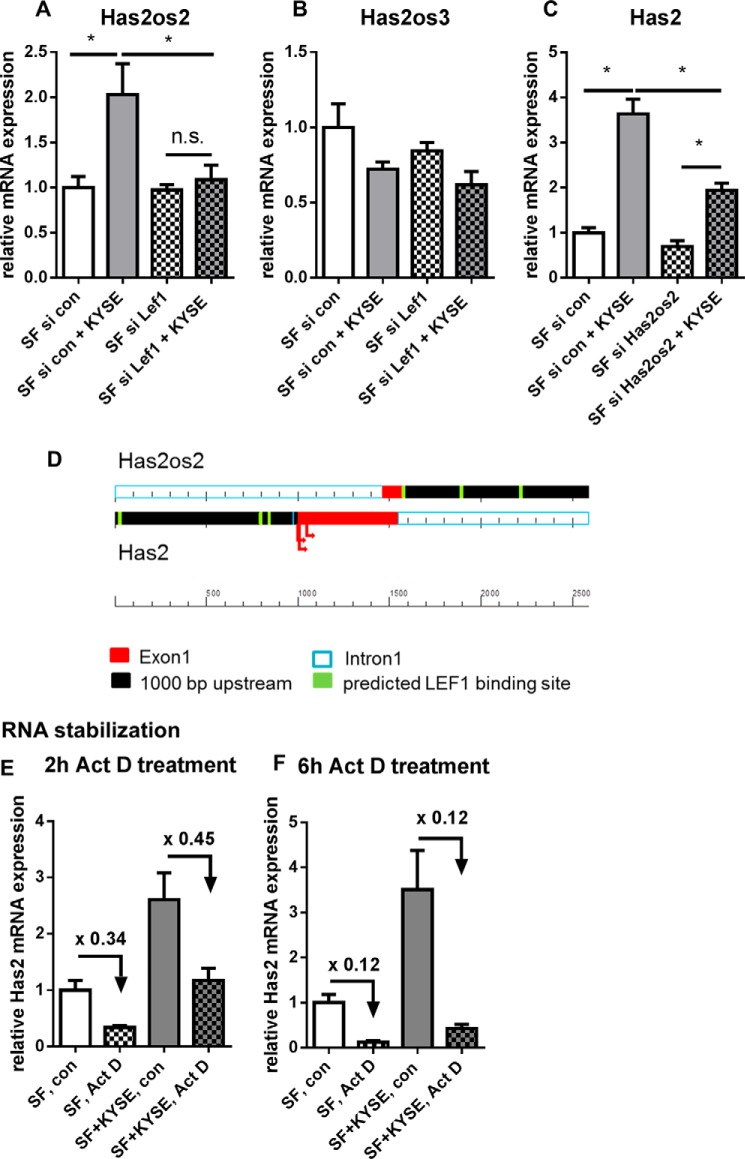
FIGURE 4.
Has2os2 RNA was regulated parallel to Has2 mRNA in co-cultures. Has2 antisense RNA Has2os1, Has2os2, and Has2os3 were measured by qPCR. Has2os1 was below the limit of detection in both monocultures and co-cultures (n = 2). A and B, Has2os2 and Has2os3 RNA expression in monocultures or direct co-cultures after transfection with siRNA directed against Lef1 or control siRNA (n = 4). C, Has2 mRNA expression in SF after knockdown of Has2os2 or control treatment (n = 4). D, scheme shows predicted LEF1 binding sites (green) in Has2 (lower strand) and Has2os2 (upper strand) promotor regions (black). Exons are depicted in red, and arrows show transcription start sites of Has2 according to the Ensembl, Uniprot, and NCBI gene databases. E and F, Has2 mRNA expression after treatment with transcriptional inhibitor actinomycin D (Act D) or vehicle control for 2 and 6 h (n = 3). Data are presented as mean ± S.E. (error bars). *, p < 0.05; n.s., not significant.