FIGURE 2.
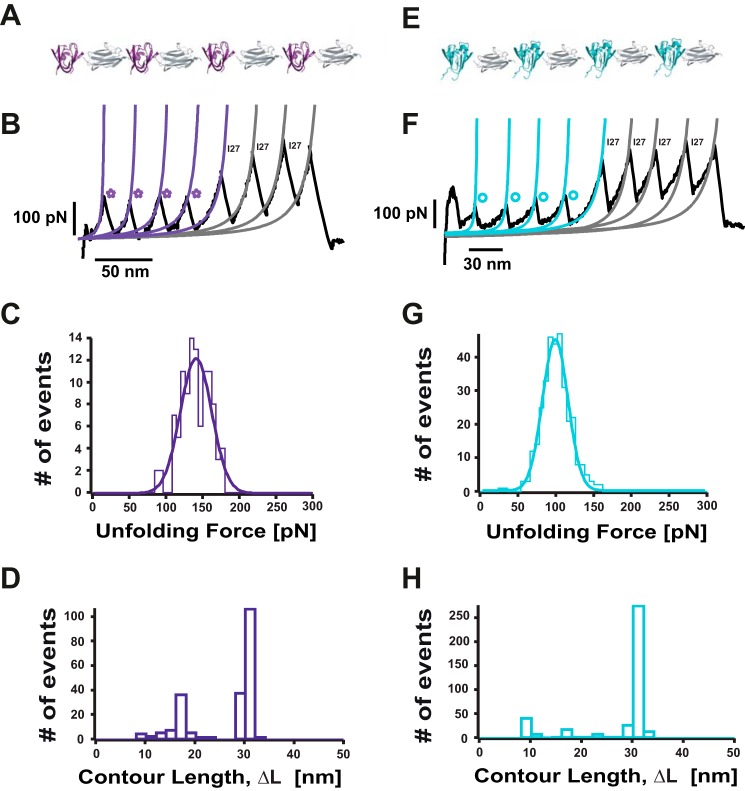
The N terminus domain of HγD-crys exhibits a higher mechanical stability than the C terminus domain. A, schematic of the (I27-Ntd)4 polyprotein. B, typical force extension unfolding trajectory of a (I27-Ntd)4 polyprotein, showing four unfolding peaks (purple stars) followed by the unfolding of the well characterized I27 protein (gray fits). C, distribution of unfolding forces for the Ntd, yielding an average unfolding force of 136.2 ± 18.0 pN (n = 106). D, distribution of the measured increment in contour length, ΔLc, upon unfolding (n = 217). Although a main unfolding ΔLc of 29.3 ± 0.7 nm is measured, other shorter ΔL values are also evident, probably because of unfolding events of partially unfolded domains. E, schematic of the (I27-Ctd)4 polyprotein. F, individual unfolding trajectory of an individual (I27-Ctd)4 polyprotein, showing the unfolding of the Ctd first (blue circles and blue Worm-like chain fits), followed by the unfolding of the I27 protein (gray Worm-like chain fits). G, mechanical unfolding of an individual Ctd requires a force of 96.1 ± 16.8 pN (n = 331). H, the distribution of associated increment in contour length ΔLc values (n = 430) shows a main unfolding ΔLc = 29.9 ± 0.8 nm together with and a much smaller population of events with associated shorter ΔLc values.