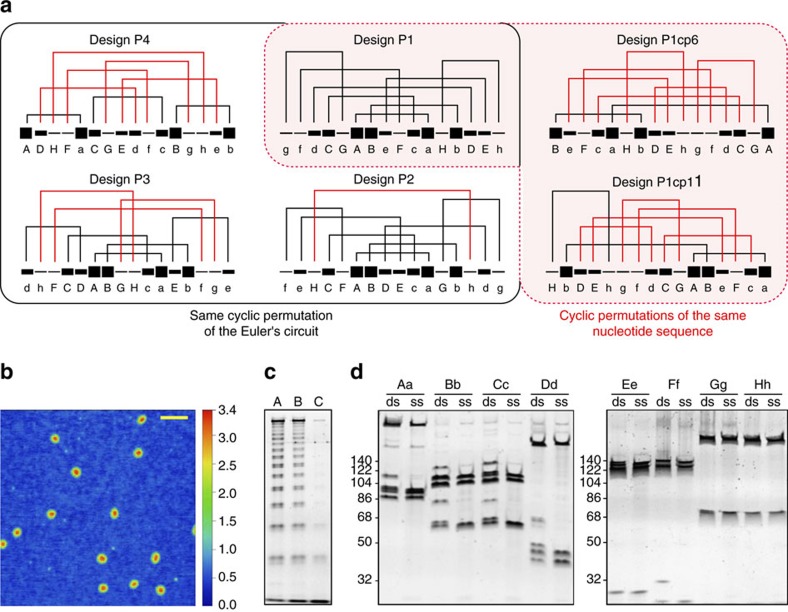
Figure 3. Single-chain DNA square pyramid designs and the structural characterization of P1.
(a) DNA square pyramid designs were constructed either by rearranging the modules within a prescribed linear topology (black box) or by a circular permutation of the polynucleotide sequence of P1 (red-shaded area). Each line segment depicts one module and is identified with a letter below. Line thickness graphically reflects the stability of complementary module pairs, the order of which was estimated based on the hPin melting curves (Supplementary Fig. 2). The stabilities are depicted on a scale from 1 to 10 and were calculated according to equation (2). Designed folding pathways (a series of connection-forming steps) are depicted for each 4 Py. Connections drawn in red correspond to topologically or kinetically frustrated steps violating the ‘free-end' rule. (b) A topography image of P1 particles deposited on mica. Scale bar, 100 nm. (c) Denaturing gradient PAGE after the Mung bean nuclease digestion of the slowly temperature annealed P1. Lines 1, 2 and 3 depict samples after 10, 100 and 300 min of digestion, respectively. (d) The denaturing gradient PAGE of P1 digestion using distinct endonucleases. Fragments produced from the digestion of end-trimmed double-stranded P1 (ds) served as controls for fragments produced after the digestion of the folded single-chain P1 (ss). Contributions from the antisense (−) strand digestion present in the control samples yet absent from the single-chain P1 containing samples result in additional bands in ds lanes. Experiments were repeated at least twice with comparable results.