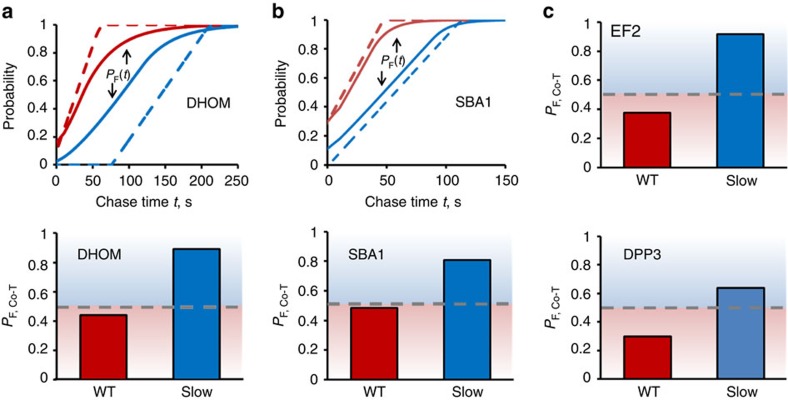
Figure 7. Synonymous codon substitutions can switch some yeast protein domains from post- to co-translational folding according to equation (2).
(a) Top panel. The probability of folding as a function of the chase time for domain 1 of DHOM predicted using equation (2). Calculations were performed for both the WT transcript (red solid line) and the transcript in which all codon positions were substituted with their slowest-translating synonymous codon (solid blue line). In the same panel is plotted the time-dependent fraction of full-length protein (see Methods section) synthesized from the WT (red dashed line) or the slow-translating (blue dashed line) transcript. (a) Bottom panel. The fraction of DHOM molecules whose first domain folds co-translationally when synthesized from the WT (red) or slowest-translating (blue) transcript. (b) Same as a but for domain 1 of SBA1. (c) Additional probabilities of co-translational folding for domain 6 of EF2 (top) and domain 2 of DPP3 (bottom) for their WT and slowest-translating transcripts. Dashed grey lines separate the co- and post-translational folding classes.