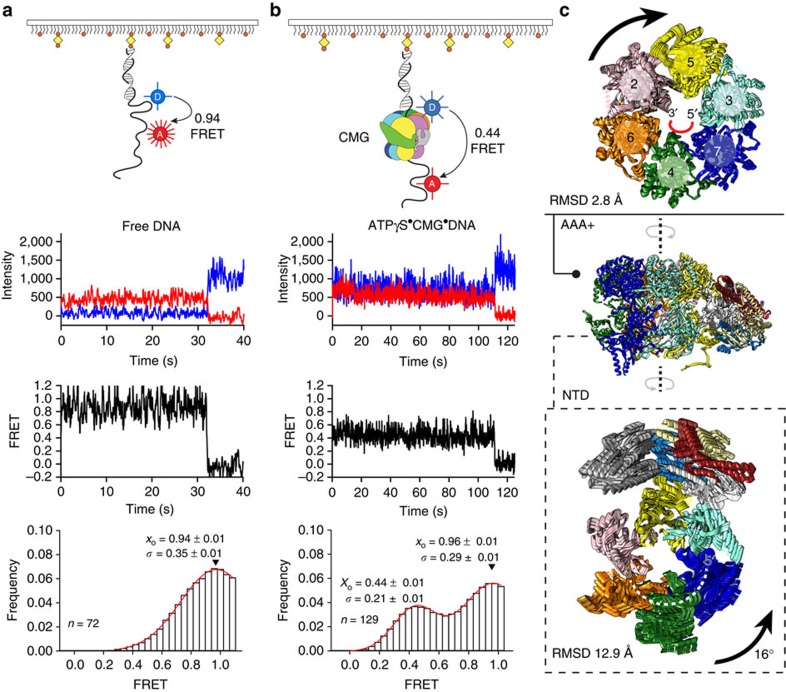
Figure 6. DNA engagement and deformation by the CMG helicase.
Single-molecule FRET analysis of (a) isolated DNA and (b) DNA+ATPγS CMG. Top to bottom: cartoon schematic of the single-molecule FRET experiment indicating FRET between Cy3 (Donor, blue sphere) and Cy5 (Acceptor, red sphere, 7-nt separation) attached to biotinylated 3′-tailed duplex DNA, immobilized on a neutravidin-coated biotin-PEG surface; donor (blue) and acceptor (red) intensity trajectories are anti-correlated until single-step photobleaching of the acceptor. FRET trajectories (black) between Cy3 and Cy5 exhibit a sharp drop to zero FRET when the acceptor photobleaches; histogram of FRET values collected from all molecules with a Gaussian fit are shown in red, where x0 is the mean FRET and σ is the distribution width. ATPase controlled ring rotation and DNA stretching by the CMG helicase. (c) The AAA+ tier of the Mcm2-7 motor rotates clockwise as Mcm2 moves towards the Mcm5 protomer, to close a gap in the motor domain. The DNA-interacting NTD rotates anticlockwise as a rigid body, together with GINS and Cdc45.