Abstract
Defining and manipulating specific neurons in the brain has garnered enormous interest in recent years, because such an approach is now widely recognized as crucial for deepening our understanding of how the brain works. When I started exploring the Cre-loxP recombination for brain research in the early 1990s, it was written off as a dead-end project by a young fool. Yet over the past 20 years, Cre-lox recombination-mediated neurogenetics has emerged as one of the most powerful and versatile technology platforms for cell-specific gene knockouts, transgenic overexpression, Brainbow imaging, neural pathway tracing with retrovirus and CLARITY, chemical genetics, and optogenetics. Its popularity and greater utility in neuroscience research is also largely thanks to the NIH’s bold Blueprint for Neuroscience Research Initiative to launch several Cre-driver resource projects, as well as individual laboratories and private research organizations. With newly-discovered, genetically-encoded molecules that are capable of responding to sonar and magnetic stimulation, for sonogenetics or magnetogenetics, respectively, or detecting rapid voltage changes in neurons, Cre-lox neurogenetics will continue to aid brain research for years to come.
Keywords: Cre-loxP, optogenetics, brainbow, mouse brain, learning and memory, neural circuits, cognition, Cre recombinase
Santiago Ramón y Cajal was among the first to observe and marvel at the elegance and beauty of various neurons in the brain and wonder how they contribute to perception, emotion, memory, and behavior (Cajal, 1909/1910). Structure-functional analysis of neurons began in earnest with the development of single-cell, juxtacellular-labeling, and patch-clamp techniques (Jankowska et al., 1976; McCrea et al., 1976; Hamill et al., 1981; Edwards et al., 1989; Pinault, 1996; Klausberger and Somogyi, 2008). Various other technologies have further transformed the neuroscience landscape at multiple levels – ranging from genes and proteins (Carraway and Leeman, 1973; Hökfelt et al., 1977; Noda et al., 1984; Seeburg et al., 1990; Buck and Axel, 1991; Moriyoshi et al., 1991; Monyer et al., 1992; Carlsson, 1993; Südhof, 2012), to synaptic plasticity and cognitive enhancement (Tang et al., 1999; Lømo, 2003; Frey and Frey, 2008; Wang et al., 2009).
In the late 1980s and early 1990s, following the pioneering efforts by Mario Capecchi, Oliver Smithies, and Martin Evans on gene targeting and embryonic stem cell (ES) technologies (see review by Capecchi, 2005), a small number of laboratories began to generate mutant mice to define genes’ function in development, cancer, or immunology (Zijlstra et al., 1989; DeChiara et al., 1990; Koller et al., 1990; Thomas and Capecchi, 1990). Alcino Silva, Seth Grant, and Thomas O’Dell were among the first to apply the gene knockout approach to study the specific roles of CaMKII or Fyn kinase in plasticity and memory (Grant et al., 1992; Silva et al., 1992a,b). These studies set a new stage to investigate genes and brain functions. However, the limitations such as the lack of phenotypes due to genetic compensation or the developmental effects were noticed. For example, increased spontaneous epilepsy was observed in the CaMKII knockout mice, whereas the dentate gyrus was deformed in the Fyn knockout mice. Both cases led to heated debate in the field about how the reported memory and plasticity deficits should be interpreted. With the NMDA receptor occupying a center stage in plasticity and memory field, Li et al. (1994) set out to demonstrate its role in learning behavior, but they found that NMDAR1 knockout pups all died neonatally due to failed brain development, including the defective suckling reflex. For developmental neuroscientists, it was very exciting, because stereotypic whisker barrel in the thalamus failed to form (Li et al., 1994). Yet for cognitive and behavioral neuroscientists, it was disappointing. It further solidified the argument that deleting the gene in every cell and organ throughout development was not ideal for defining cognitive mechanisms.
Young and Foolish
My interest in developing the conditional gene knockout technique stemmed from the specific problem at my hand as I was finalizing my HHMI postdoctoral fellowship with Eric Kandel at Columbia University in the summer of 1993. From 1990 to 1993, I was testing the long-held idea, first postulated by Bernie Agranoff (Agranoff and Klinger, 1964), that long-term memory requires new protein synthesis and gene expression. Researchers were racing to identify novel genes whose expressions were regulated by brain activity. We were the first to pull out a set of genes which included tissue-plasminogen activator (tPA), a MAP kinase phosphatase, and a brain-specific immediate early gene BAD1 (published under my former name, Qian et al., 1993, 1994). BAD1 was later also isolated by Paul Worley and named as Arc (Lyford et al., 1995). While isolating these novel genes was exciting, the next logical step for me was to examine their functions in memory. Anti-sense oligonucleotide-based “knock-down” methods were imprecise and not reliable, whereas gene-knockout was a good choice, but its limitations and caveats were obvious.
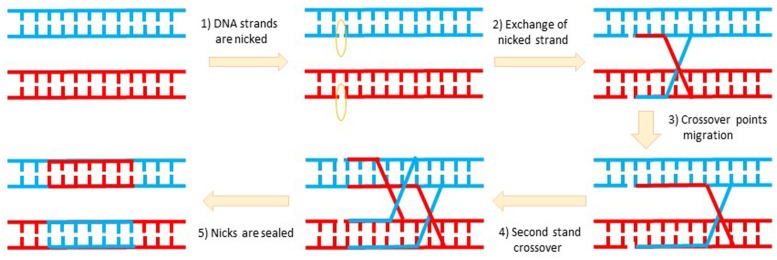
I stumbled upon a paper by Brian Sauer on his successful demonstration of the Cre recombinase-based excision of a floxed marker gene from the circular plasmid transfected in mammalian culture cells (Sauer and Henderson, 1988). The last paragraph of this paper put forth a big question: “Can Cre also cause recombination at lox sites located within the genome of a mammalian cell?” This led me to wonder if I could use it to develop brain subregion- and cell type-specific gene knockout and/or transgenic overexpression methods. But I knew that all the textbooks and literature describing DNA replication and recombination go hand in hand with cells division (such as meiosis or mitosis; Figure 1). This basic doctrine was imprinted into everyone’s mind, and also evident from the opening sentences of Sauer’s paper: “The processes governing DNA recombination in mitotic mammalian cells have been the subject of intense investigation in recent years… Mitotic recombination plays a central role in the development and function of the immune system.” Knowing that all neurons in the adult brain are known to be postmitotic right after birth (except a few in the dentate gyrus and olfactory bulb), any fool who set out to work on DNA recombination in the adult brain would be committing a career suicide. Nature has shown that brain tumors are all in glial cells which divide, but not in neurons. Therefore, DNA recombination as the way to create region- and cell type-specific knockout in the brain was considered to be plainly impossible. I was told that the next show was not about how and why I would fail, but where.
FIGURE 1.
DNA recombination involves complex topological and biochemical during DNA replication. DNA recombination is linked with DNA replication during cell divisions in order to exchange genetic materials for the selection of favorable mutations or the elimination of unfavorable or deleterious mutations. Recombination also comes with DNA injury and is therefore tightly coupled with the DNA repair mechanism. Nearly all neurons in the adult brain are post-mitotic to ensure their stability. This explains why brain tumors are in glial cells but not in neurons.
But I was obsessed with the thought of wanting of know the function of genes in a clear way, and only a handful of laboratories had gene-targeting facilities and embryonic stem (ES) cells for making conventional knockout mice. I asked Mario Capecchi of Utah and Susumu Tonegawa of MIT for a second postdoc position, and both said yes. When I sought advice, to my great surprise, Eric suggested that I should choose Susumu’s lab as it would be a better place for trying my idea. I also got the permission to use the CaMKII promoter, cloned by Mark Mayford, which becomes active after the second or third postnatal week and only expresses the forebrain principal cells such as pyramidal cells (Mayford et al., 2005). The stage was all set, with the only minor bug: if my idea did not work out at this second postdoc stage, I would be out of a job.
When I arrived at MIT in the fall of 1993, I was pleased to find that Susumu did not discourage me from exploring Cre-loxP neurogenetics during our 15-s introduction meeting. I was surprised by the zoo-like atmosphere in this 40 postdoctorals/students laboratory. Miraculously, this survival-of-the-fittest model worked well for productivity, and overall MIT was exciting and refreshing to me.
To develop Cre-loxP-neurogenetics, I must ignore three risks: (1) to hope the textbook on DNA recombination linked with replication was wrong, and my argument was that herpes virus (producing cold sore) infected the peripheral nerve ganglia and somehow replicated itself. (The mechanism still remains unknown to date, but presumably without ganglia neuron division.) (2) The complex procedures and long cycles with no quick feedback and no room for error. I must make various constructs and generate at least three different mouse lines and then embark on a multi-year crossing to breed them together (Figures 2A,B). Back then, making even one mutant mouse was already a form of art and luck. In fact, only a few brave souls in well-equipped laboratories could do so because of the complex procedures, lengthy project cycle, expensive cost, and with potentially no phenotypes or undesirable outcomes at the end. A few lucky ones got good jobs and the other unlucky ones simply disappeared. (3) The intentional risk which I brought upon myself: I chose to work on the NMDAR1 as the gene for conditional knockout, instead of the BAD1/Arc. As everyone knew, if the sites to which I inserted the LoxP somehow disrupted its expression, I would have a dead pup that was to be published in a few months by Li et al. (1994). In contrast, unintentional disruption of Arc from the loxP insertion would still give me a conventional knockout paper. But I convinced myself the idea was promising, and my gamble would pay off: I would, at least, get a badge of honor for being the first fool to throw myself under the bus of the textbook dogma.
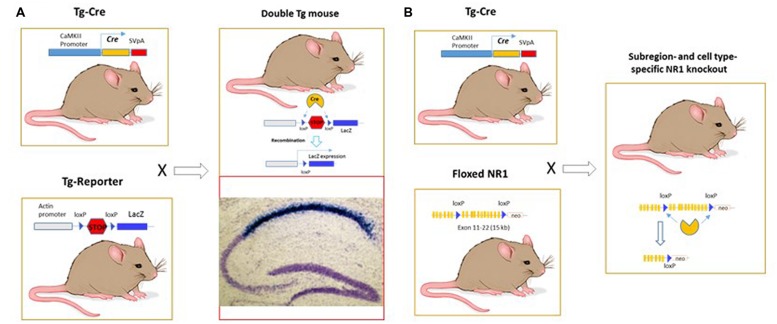
FIGURE 2.
Cre-LoxP neurogenetics for achieving region- and cell-type specific analysis of relationships of genes, circuits, and functions in the brain. (A) Strategy to detect the Cre-LoxP recombination in the brain. It required the production of two different transgenic mice: Tg-Cre and Tg-Reporter lines, respectively. These two lines were then crossed to generate the double transgenic mice. CA1-specific recombination was detected after P19 by LacZ method. It is known that CA1 pyramidal cells undergo neurogenesis between E10 and E18 and enter the post-mitotic state by P0. They are well differentiated by P7, with fully established synaptic connections. We have found that Cre-loxP recombination occurs during the middle or end of the third postnatal week in the CA1 pyramidal cells. LacZ in the CA1 pyramidal cells was detected as deep blue color on Nissl stain (in purple-blue) background. (B) Conditional knockout of NR1 gene in a specific cell type and region. The exon 11-21 encoding the entire transmembrane domain and C-terminus were flanked by the loxP (termed floxed) in the introns. The second loxP was followed with the neo cassette gene, which allowed for targeted ES cells selection. Luckily, insertion of loxP sites and the neo gene did not alter NR1 gene expression in the floxed homozygous mice.
The execution required complex choreography. Despite all of the chaos in the new laboratory, I found friendly colleagues willing to help me. There was David Gerber and Toshikuni Sasaoka who taught me about microinjections in fertilized eggs and blastocysts, respectively; Dongfeng Chen who helped with immune-antibody and staining; Yuqing Li who shared his insights into the NMDAR1 constructs; Min Xu who provided me ES cells and guided me through delicate ES cell-culture procedures; I was also grateful to David Anderson at CalTech for the LacZ reporter and Brian Saucer at Du Pont for Cre-loxP plasmids. I completed all my constructs by January of 1994 and started production of transgenic mice and ES cell targeting of the floxed NR1 construct. I also had a hard-working undergraduate student named Cindy Tom and technician Jason Derwon, who assisted with the genotyping and brain sectioning.
One ominous cloud moved over my head when Klaus Rajewsky reported in July 1994 that they achieved 50% knocked down of the RNA polymerase in T cells (Gu et al., 1994). This meant that even in dividing T cells, Cre-recombination was not efficient: either 50% of the T cells had 100% knockout of RNA polymerase or 100% of the cells had only one copy of the gene deleted, or worse, a mixed mosaic situation. Despite this, I held out the hope (after additional digging into the literature) that the transiently active promoter they used might be the culprit for the poor efficiency during T cell development. Over the ensuing months and years, I stuck my head in the sand and labored over my Cre-loxP neurogenetics experiments. In the late fall of 1994, I finally got the first feedback from the experiments.
On a sunny but cold morning, I recall the great surprise when I saw the intense LacZ staining, specifically in the CA1 pyramidal cells of the hippocampus in the first Cre transgenic line (Figure 2). I could not believe my incredible luck, because the CA1 hippocampal region was the center of the universe in the eyes of many plasticity and memory researchers. Additional Cre lines confirmed similar CA1-specific recombination; then other Cre lines showed forebrain-specific patterns. When Susumu returned from a trip to Japan, I told him about what I had found. Once he recovered from his confusion, which seemed to result from jetlag and attempting to grasp all that I had been working on, he overcame his disbelief and immediately shared his jubilation with Alcino over the phone. I asked myself why the CaMKII promoter, which was supposed to express in the forebrain – not specific to the CA1- had such a specific effect. Dongfeng Chen helped provide the clue by revealing that Cre expression was higher in the CA1 pyramidal cells. Over the following 6–8 months, I also obtained the floxed NR1 homozygous mice crossed with the Cre Tg 29.1 line. I confirmed the CA1 pyramidal cell-specific NMDA receptor knockout. Pato Huerta and I proceeded with brain slice recordings or histology, respectively, as soon as we could obtain young adult mice (∼3–5 weeks). We also set aside a set of mice for collaborating with Tom McHugh, Kenny Blum, and Matthew Wilson for place cell recordings in the NR1 mutants. Now, we know that the Cre-lox recombination in T29-1 retained the CA1 specificity only in an age-dependent manner: as Cre expression accumulated over time (starting in the 6th–8th postnatal weeks), it would gradually achieve the forebrain-specific recombination (as expected from the CaMKII promoter).
By the fall of 1996, we were preparing three back-to-back manuscripts to be submitted to Cell. We had decided that reporting the feasibility of the Cre-lox neurogenetics would be the first, the CA1-specific NMDA receptor knockout study the second, and the place-cell characterization paper would be published as the third. The original agreement with Eric for using the CaMKII promotor was to include him and Mark as the co-authors in the first paper, in case the Cre-lox work progressed to a point of publication. This initially doomed project now unexpectedly resulted in multiple results, which created its own political nuisance. Knowing the immense interest of the CA1-specific NMDA receptor knockouts in the learning and memory field, Eric preferred his co-authorship be placed in the second paper, not the first Methodology paper. But Susumu insisted that Eric’s name be in the first paper. The back-and-forth conversations that ensued were very distressful to me, but allowed my brain to form an unusually strong long-term semantic memory of what it means to be caught between a rock and a hard place. The papers were eventually published in December 1996 (McHugh et al., 1996; Tsien et al., 1996a,b), but they were left with no corresponding author.
Cre-Driver Resources for Neurosciences
The successful demonstration of the feasibility of Cre-lox neurogenectis has generated a firestorm in the field. Recognizing its unique usefulness for neuroscience, the NIH launched the NIH Blueprint for Neuroscience Research, known as the Cre-driver project, to create a collection of mouse strains for better defining the functions of specific cell types and neural circuits in cognitive behaviors1. I was glad to provide my inputs during the process. The NIH chose three centers in the United States for the generation of genetically modified mice expressing Cre recombinases in the nervous system. The teams were led by Dr. Ronald Davis at Baylor College of Medicine (now at Scripps Research Institute-Florida), Dr. Josh Huang at Cold Spring Harbor Laboratory (co-led with Sacha Nelson of Brandeis University) and Dr. Ulrich Mueller at Scripps Research Institute. This program generated more than hundreds of Cre-driver lines (Taniguchi et al., 2011). The mice are available through the Mutant Mouse Regional Resource Center (MMRRC) or the Jackson Laboratory’s Cre Repository (see Table 1).
Table 1.
Cre-driver mouse resources.
| Cre-driver mice Network | NIH Blueprint for neuroscience | http://www.credrivermice.org |
| MMRRC (Mutant Mouse Regional Resource Center) | https://www.mmrrc.org/cataloR/StrainCatalogSearchForm.isp | |
| The Jackson Laboratory’s Cre Repository | https://www.jax.org/jax-mice-and-services | |
| The GENSAT project | NIH Blueprint/Rockefeller University | http://www.gensat.org/cre.isp |
| EMMA (European Mouse Mutant Archive) | European Union, Italy. | www.emmanet.org |
| MSR (International Mouse Strain Resource) | www.findmice.org | |
| Harwell (Mammalian Genetics Unit) | UK | www.har.mrc.ac.uk |
| Cre-X-mice Project | Canada | http://nagy.mshri.on.ca/cre_new/index.php |
| RBRC (RIKEN Biology Resource Center) | Japan | http://mus.brc.riken.jp/en/ |
The NIH Neuroscience Blueprint program also funded the GENSAT project led by Nathaniel Heintz of The Rockefeller University to generate BAC-Cre recombinase driver mouse lines (Gong et al., 2007). A total of 288 Cre lines have been generated to date2. Following these efforts, many individual laboratories and institutions also produced a variety of Cre drivers (e.g., Madisen et al., 2010, 2015; Leão et al., 2012). Altogether, at least over 630 Cre lines have been deposited at the Jackson Laboratory to date.
The European Union (EU) also followed the NIH initiative and launched its own Cre-driver project under the name CREATE (Coordination of resources for conditional expression of mutated mouse alleles) (Table 1). The CREATE consortium represents a core of major European and international mouse database holders and research groups to develop a strategy for the integration and dissemination of Cre-driver strains for modeling aspects of complex human diseases in the mouse. These Cre lines can be found by contacting EMMA (European Mouse Mutant Archive, Italy) or MSR (International Mouse Strain Resource; Table 1). In addition, the United Kingdom, Canada, and Japan have also funded several Cre-driver projects (Table 1).
Cre-Driving Toward Multiple Directions into the Future
In the past decade, one of the most exciting developments is optogenetics which uses light stimulation to manipulate neurons (Zemelman et al., 2002; Boyden et al., 2005). It has been great fun to witness that optogenetics was able to take full advantage of a decade’s investment in generating a rich collection of Cre mouse lines (e.g., CaMKII::Cre, PV::Cre, SOM::Cre, D1: Cre, D2::Cre, ChAT:Cre, or DAT::Cre, etc.), enabling its widespread use in the neuroscience community (Madisen et al., 2012).
However, optogenetics was not the only game in town that benefited from Cre-loxP neurogenetics. Chemical genetics has also emerged as a powerful approach in allowing scientists to activate or suppress specific neurons. For example, one can over-express chemically–genetically modified proteins, such as kinases (Bishop et al., 2000; Wang et al., 2003), or DREADD (Rogan and Roth, 2011) in specific neurons or brain regions. Researchers can also use the floxed diphtheria toxin fragment A (DTA) to delete specific cells using cre lines (Brockschnieder et al., 2004; Matsumura et al., 2004; Buch et al., 2005; Ivanova et al., 2005), and then observe the resulting phenotypes.
In addition, Cre-lox neurogenetics are used to track neural projections in the brain with retrograde virus (Sun et al., 2014; Beier et al., 2015) or CLARITY methods (Lerner et al., 2015). Jeff Lichtman and Joshua Sanes have cleverly explored the unique properties of the Cre-loxP system to randomly express different ratios of red, green, and blue derivatives of green fluorescent protein (GFP) in individual neurons, a technique termed Brainbow (Livet et al., 2007). This allowed them to flag each neuron with a distinctive color. This process has been a major contribution to the field of connectomics.
Cre-lox neurogenetics has also been combined with voltage-sensitive proteins to monitor changes in neuronal activity. Genetically encoded calcium indicators, such as the GCaMPs, have allowed researchers to infer changes in neuron activations via calcium transients (Tian et al., 2009). Researchers are also developing other genetically encoded voltage-sensitive fluorescent proteins that can report cortical electrical responses in vitro and/or in vivo (Akemann et al., 2010; Ghitani et al., 2015; Madisen et al., 2015).
Most recently, Stuart Ibsen and Sreekanth Chalasani reported an exciting way to use low-pressure ultrasound stimulation to produce microbubbles that amplify the mechanical deformations which can be detected by TRP-4, the pore-forming subunit of a mechanotransduction channel (Ibsen et al., 2015). By overexpressing TRP-4 in specific neurons, the authors showed it can sensitize neurons to an ultrasound stimulus. Similarly exciting, Sheng-Jia Zhang and his colleagues recently reported another noninvasive method, namely magnetogenetics, to activate neurons in vivo using magnetic stimulation. The activation of neurons was achieved by neuronal expression of an exogenous magnetoreceptor, an iron–sulfur cluster assembly protein 1 (Isca1) (Long et al., 2015). These two noninvasive approaches offer additional tools to manipulate specific neurons in behaving animals.
Looking back, Cre-lox neurogenetics was written off as a dead-end project, but my intuition and diligence got me through the door. Nowadays, a week rarely goes by without having yet another Cre-based neuroscience finding reported. Cre-lox neurogenetics seems to still have a lot of miles ahead of it. To my students, I always say that being young and foolish can be an asset for the community!
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Georgia Research Alliance and National Institutes of Health (R01NS079774). I also would like to express my gratitude for the unique opportunity of working at the Brain Decoding Project Consortium organized by Banna Biomedical Research Institute and supported by Yunnan Science Commission (2014DG002). I am also grateful to the Frontiers journal and editors for their kind invitation to contribute to this special issue.
Footnotes
References
- Agranoff B. W., Klinger P. D. (1964). Puromycin effect on memory fixation in the goldfish. Science 146 952–953. 10.1126/science.146.3646.952 [DOI] [PubMed] [Google Scholar]
- Akemann W., Mutoh H., Perron K., Rossier J., Knöpfel T. (2010). Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat. Methods 7 643–649. 10.1038/nmeth.1479 [DOI] [PubMed] [Google Scholar]
- Beier K. T., Steinberg E. E., DeLoach K. E., Xie S., Miyamichi K., Schwarz L., et al. (2015). Circuit architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell 162 622–634. 10.1016/j.cell.2015.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., et al. (2000). A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407 395–401. 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8 1263–1268. 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- Brockschnieder D., Lappe-Siefke C., Goebbels S., Boesl M. R., Nave K. A., Riethmacher D. (2004). Cell depletion due to diphtheria toxin fragment a after Cre-mediated recombination. Mol. Cell. Biol. 24 7636–7642. 10.1128/MCB.24.17.7636-7642.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T., Heppner F. L., Tertilt C., Heinen T. J., Kremer M., Wunderlich F. T., et al. (2005). Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2 419–426. 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 1991 175–187. 10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- Cajal S. R. (1909/1910). Histology of the Nervous System (trans. Swanson N., Swanson L. W.). New York, NY: Oxford University Press. [Google Scholar]
- Capecchi M. R. (2005). Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6 507–512. 10.1038/nrg1619 [DOI] [PubMed] [Google Scholar]
- Carlsson A. (1993). Thirty years of dopamine research. Adv. Neurol. 60 1–10. [PubMed] [Google Scholar]
- Carraway R., Leeman S. E. (1973). The isolation of a new hypotensive peptide, neurotensin, from bovine hypothalami. J. Biol. Chem. 248 6854–6861. [PubMed] [Google Scholar]
- DeChiara T. M., Efstratiadis A., Robertson E. J. (1990). A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345 78–80. 10.1038/345078a0 [DOI] [PubMed] [Google Scholar]
- Edwards F. A., Konnerth A., Sakmann B., Takahashi T. (1989). A thin slice preparation for patch clamp recordings from neurones of the mammalian central nervous system. Pflugers Arch. 414 600–612. 10.1007/BF00580998 [DOI] [PubMed] [Google Scholar]
- Frey S., Frey J. U. (2008). ‘Synaptic tagging’ and ‘cross-tagging’ and related associative reinforcement processes of functional plasticity as the cellular basis for memory formation. Prog. Brain Res. 169 117–143. 10.1016/S0079-6123(07)00007-6 [DOI] [PubMed] [Google Scholar]
- Ghitani N., Bayguinov P. O., Ma Y., Jackson M. B. (2015). Single-trial imaging of spikes and synaptic potentials in single neurons in brain slices with genetically encoded hybrid voltage sensor. J. Neurophysiol. 2015 1249–1259. 10.1152/jn.00691.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., Doughty M., Harbaugh C. R., Cummins A., Hatten M. E., Heintz N., et al. (2007). Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci. 27 9817–9823. 10.1523/JNEUROSCI.2707-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. G., O’Dell T. J., Karl K. A., Stein P. L., Soriano P., Kandel E. R. (1992). Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science 258 1903–1910. 10.1126/science.1361685 [DOI] [PubMed] [Google Scholar]
- Gu M., Marth J. D., Orban P. C., Mossmann H., Rajewsky K. (1994). Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science 265 103–106. 10.1126/science.8016642 [DOI] [PubMed] [Google Scholar]
- Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981). Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 391 85–100. 10.1007/BF00656997 [DOI] [PubMed] [Google Scholar]
- Hökfelt T., Elfvin L. G., Elde R., Schultzberg M., Goldstein M., Luft R., et al. (1977). Occurrence of somatostatin-like immunoreactivity in some peripheral sympathetic noradrenergic neurons. Proc. Natl. Acad. Sci. U.S.A. 74 3587–3591. 10.1073/pnas.74.8.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen S., Tong A., Schutt C., Esener S., Chalasani S. H. (2015). Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat. Commun. 6:8264 10.1038/ncomms9264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A., Signore M., Caro N., Greene N. D. E., Copp A. J., Martinez-Barbera J. P. (2005). In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment a. Genesis 43 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska E., Rastad J., Westman J. (1976). Intracellular application of horseradish peroxidase and its light and electron microscopical appearance in spinocervical tract cells. Brain Res. 105 557–562. 10.1016/0006-8993(76)90603-X [DOI] [PubMed] [Google Scholar]
- Klausberger T., Somogyi P. (2008). Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 4 53–57. 10.1126/science.1149381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller B. H., Marrack P., Kappler J. W., Smithies O. (1990). Normal development of mice deficient in β2M, MHC class I. proteins, and CD8+ T cells. Science 248 1227–1230. 10.1126/science.2112266 [DOI] [PubMed] [Google Scholar]
- Leão R. N., Mikulovic S., Leão K. E., Munguba H., Gezelius H., Enjin A., et al. (2012). OLM interneurons differentially modulate CA3 and entorhinal inputs to hippocampal CA1 neurons. Nat. Neurosci. 15 1524–1530. 10.1038/nn.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner T. N., Shilyansky C., Davidson T. J., Evans K. E., Beier K. T., Zalocusky K. A., et al. (2015). Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162 635–647. 10.1016/j.cell.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Erzurumlu R. S., Chen C., Jhaveri S., Tonegawa S. (1994). Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell 76 427–437. 10.1016/0092-8674(94)90108-2 [DOI] [PubMed] [Google Scholar]
- Livet J., Weissman T. A., Kang H., Draft R. W., Lu J., Bennis R. A., et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450 56–62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Lømo T. (2003). The discovery of long-term potentiation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 617–620. 10.1098/rstb.2002.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Ye J., Zhao D., Zhang S.-J. (2015). Magnetogenetics: remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Sci. Bull. 60 2107–2119. 10.1007/s11434-015-0902-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford G. L., Yamagata K., Kaufmann W. E., Barnes C. A., Sanders L. K., Copeland N. G., et al. (1995). Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14 433–445. 10.1016/0896-6273(95)90299-6 [DOI] [PubMed] [Google Scholar]
- Madisen L., Garner A. R., Shimaoka D., Chuong A. S., Klapoetke N. C., Li L., et al. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85 942–958. 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Mao T., Koch H., Zhuo J. M., Berenyi A., Fujisawa S., et al. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15 793–802. 10.1038/nn.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H., Hasuwa H., Inoue N., Ikawa M., Okabe M. (2004). Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin a chain. Biochem. Biophys. Res. Commun. 321 275–279. 10.1016/j.bbrc.2004.06.139 [DOI] [PubMed] [Google Scholar]
- Mayford M., Wang J., Kandel E. R., O’Dell T. J. (2005). CaMKII regulates the frequency-response function of hippocampal synapses for the production of both LTD and LTP. Cell 81 891–904. 10.1016/0092-8674(95)90009-8 [DOI] [PubMed] [Google Scholar]
- McCrea R. A., Bishop G. A., Kitai S. T. (1976). Intracellular staining of purkinje cells and their axons with horseradish peroxidase. Brain Res. 118 132–136. 10.1016/0006-8993(76)90847-7 [DOI] [PubMed] [Google Scholar]
- McHugh T. J., Blum K. I., Tsien J. Z., Tonegawa S., Wilson M. A. (1996). Impaired hippocampal representation of space in CA1-specific NMDAR1 knockout mice. Cell 87 1339–1349. 10.1016/S0092-8674(00)81828-0 [DOI] [PubMed] [Google Scholar]
- Monyer H., Sprengel R., Schoepfer R., Herb A., Higuchi M., Lomeli H., et al. (1992). Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256 1217–1221. 10.1126/science.256.5060.1217 [DOI] [PubMed] [Google Scholar]
- Moriyoshi K., Masu M., Ishii T., Shigemoto R., Mizuno N., Nakanishi S. (1991). Molecular cloning and characterization of the rat NMDA receptor. Nature 354 31–37. 10.1038/354031a0 [DOI] [PubMed] [Google Scholar]
- Noda M., Shimizu S., Tanabe T., Takai T., Kayano T., Ikeda T., et al. (1984). Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature 312 121–127. 10.1038/312121a0 [DOI] [PubMed] [Google Scholar]
- Pinault D. (1996). A novel single-cell staining procedure performed in vivo under electrophysiological control: morpho-functional features of juxtacellularly labeled thalamic cells and other central neurons with biocytin or Neurobiotin. J. Neurosci. Methods 65 113–136. 10.1016/0165-0270(95)00144-1 [DOI] [PubMed] [Google Scholar]
- Qian Z., Gilbert M., Kandel E. R. (1994). Temporal and spatial regulation of the expression of BAD2, a MAP kinase phosphatase, during seizure, kindling, and long-term potentiation. Learn. Mem. 1 180–188. 10.1101/lm.1.3.180 [DOI] [PubMed] [Google Scholar]
- Qian Z., Tsien J. Z., Gilbert M. E., Colicos M. A., Kandel E. R., Kuhl D. (1993). Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature 361 453–457. 10.1038/361453a0 [DOI] [PubMed] [Google Scholar]
- Rogan S. C., Roth B. L. (2011). Remote control of neuronal signaling. Pharmacol. Rev. 63 291–315. 10.1124/pr.110.003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., Henderson N. (1988). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Nat. Acad. Sci. U.S.A. 85 5166–5170. 10.1073/pnas.85.14.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg P. H., Wisden W., Verdoorn T. A., Pritchett D. B., Werner P., Herb A., et al. (1990). The GABAA receptor family: molecular and functional diversity. Cold Spring Harb. Symp. Quant. Biol. 55 29–40. 10.1101/SQB.1990.055.01.006 [DOI] [PubMed] [Google Scholar]
- Silva A. J., Stevens C. F., Tonegawa S., Wang Y. (1992a). Deficient hippocampal long-term potentiation in α-calcium-calmodulin kinase II mutant mice. Science 257 201–206. 10.1126/science.1321493 [DOI] [PubMed] [Google Scholar]
- Silva A. J., Paylor R., Wehner J. M., Tonegawa S. (1992b). Impaired spatial learning in α-calcium-calmodulin kinase II mutant mice. Science 257 206–211. 10.1126/science.1321493 [DOI] [PubMed] [Google Scholar]
- Südhof T. C. (2012). Calcium control of neurotransmitter release. Cold Spring Harb. Perspect. Biol. 4:a011353 10.1101/cshperspect.a011353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nguyen A. Q., Nguyen J. P., Le L., Saur D., Choi J., et al. (2014). Cell-type-specific circuit connectivity of hippocampal CA1 revealed through Cre-dependent rabies tracing. Cell Rep. 7 269–280. 10.1016/j.celrep.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. P., Shimizu E., Dube G. R., Rampon C., Kerchner G. A., Zhuo M., et al. (1999). Genetic enhancement of learning and memory in mice. Nature 401 63–69. 10.1038/43432 [DOI] [PubMed] [Google Scholar]
- Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71 995–1013. 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. (1990). Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 346 847–850. 10.1038/346847a0 [DOI] [PubMed] [Google Scholar]
- Tian L., Hires S. A., Mao T., Huber D., Chiappe M. E., Chalasani S. H., et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6 875–881. 10.1038/nmeth.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien J. Z., Chen D. F., Gerber D., Tom C., Mercer E. H., Anderson D. J., et al. (1996a). Subregion- and cell type-restricted gene knockout in mouse brain. Cell 87 1317–1326. 10.1016/S0092-8674(00)81826-7 [DOI] [PubMed] [Google Scholar]
- Tsien J. Z., Huerta P. T., Tonegawa S. (1996b). The essential role of hippocampal CA1 NMDA receptor–dependent synaptic plasticity in spatial memory. Cell 87 1327–1338. 10.1016/S0092-8674(00)81827-9 [DOI] [PubMed] [Google Scholar]
- Wang D., Cui Z., Zeng Q., Kuang H., Wang L. P., Tsien J. Z., et al. (2009). Genetic enhancement of memory and long-term potentiation but not CA1 long-term depression in NR2B transgenic rats. PLoS ONE 4:e7486 10.1371/journal.pone.0007486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Shimizu E., Tang Y. P., Cho M., Kyin M., Zuo W., et al. (2003). Inducible protein knockout reveals temporal requirement of CaMKII reactivation for memory consolidation in the brain. Proc. Natl. Acad. Sci. U.S.A. 100 4287–4292. 10.1073/pnas.0636870100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemelman B. V., Lee G. A., Ng M., Miesenböck G. (2002). Selective photostimulation of genetically chARGed neurons. Neuron 33 15–22. 10.1016/S0896-6273(01)00574-8 [DOI] [PubMed] [Google Scholar]
- Zijlstra M., Li E., Sajjadi F., Subramani S., Jaenisch R. (1989). Germ-line transmission of a disrupted β2-microglobulin gene produced by homologous recombination in embryonic stem cells. Nature 342 435–438. 10.1038/342435a0 [DOI] [PubMed] [Google Scholar]