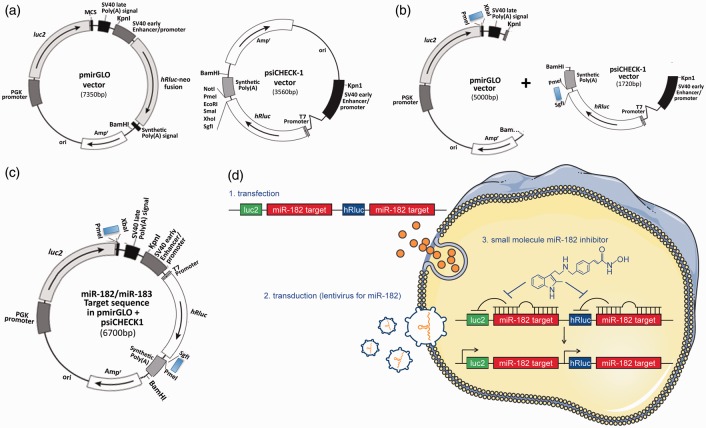
Figure 1.
Generation of dual-luciferase miRNA target expression constructs and their stable transfectants. (a) Commercially available original vectors, pmirGLO and psiCHECK-1, which were designed to quantitatively evaluate miRNA activity via the insertion of miRNA target sites on the 3′ UTR of the firefly gene (luc2) (pmirGLO) or 3′ UTR of the Renilla gene (hRluc) (psiCHECK-1). (b) Insertion of annealed oligonucleotide pairs, which contain the miR-182 (or miR-183) target sequence with appropriate restriction sites (PmeI and XbaI for pmirGLO, SgfI and PmeI for psiCHECK-1) downstream of luc2 in the pmirGLO and downstream of hRluc in the psiCHECK-1. These vectors were digested with KpnI and BamHI and the fragments which contained the reporter units were isolated/ligated. (c) The final construct consists of a dual reporter system utilizing both firefly and Renilla luciferase. (d) (1) Using SHSY5Y cells, stable transfectants of the engineered reporter constructs were created (specific for either miR-182 [represented] or miR-183). (2) In order to maintain minimal basal levels of activity of both luciferases, the stable host cells were transduced with lentiviral particles containing miR-182 (or miR-183) shMIMIC microRNAs. (3) These stable transfectants (miR-182 or miR-183 target sequence in pmirGLO/psiCHECK1 plus lentiviral particles containing miR-182 or miR-183 shMIMIC) were usable for high-throughput screens. The final stable reporter cell line was designed to identify small compounds (e.g. HDAC inhibitor Panobinostat) that inhibit miR-182 (and/or miR-183) generation or function, thereby resulting in the activation of the luciferases (both firefly and Renilla).