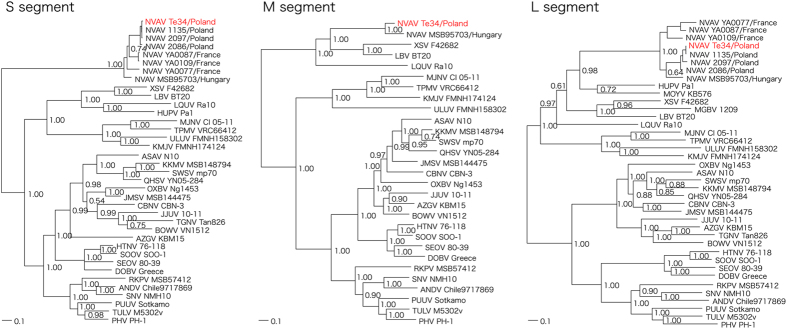
Figure 2. Phylogenetic trees generated by the Bayesian method, using the GTR+I+Γ model of evolution as estimated from the data, based on the alignment of the coding regions of the full- length S, M and L segments of NVAV strain Te34.
Topologies of the unrooted phylogenetic trees using the maximum-likelihood method were nearly identical. Relationships are shown to the prototype NVAV strain from Hungary (MSB95703, S: FJ539168; M: HQ840957; L: FJ593498) and to representative NVAV strains from Poland (1135, S: JX990924; L: JX990948; 2086, S: KF515970; L: JX990963; and 2097, S: JX990930; L: KF663727) and France (YA0077, S: KF010573; L: KF010538; YA0087, S: KF010571; L: KF010534; YA0109, S: KF010565; L: KF010521). Other hantaviruses harbored by shrews and moles included Thottapalayam virus (TPMV VRC66412), Imjin virus (MJNV Cl 05-11), Uluguru virus (ULUV FMNH158302), Kilimanjaro virus (KMJV FMNH174124), Asama virus (ASAV N10), Oxbow virus (OXBV Ng1453), Rockport virus (RKPV MSB57412), Jemez Springs virus (JMSV MSB144475), Seewis virus (SWSV mp70), Kenkeme virus (KKMV MSB148794), Qian Hu Shan virus (QHSV YN05-284), Cao Bang virus (CBNV CBN-3), Azagny virus (AZGV KBM15), Tanganya virus (TGNV Tan826), Bowé virus (BOWV VN1512), and Jeju virus (JJUV SH42). Bat-borne hantaviruses included Magboi virus (MGBV MGB1209), Mouyassué virus (MOYV KB576), Huangpi virus (HUPV Pa-1), Longquan virus (LQUV Ra-10), Laibin virus (LBV BT20), and Xuan Son virus (XSV F42682). Also shown are representative rodent-borne hantaviruses, including Hantaan virus (HTNV 76-118), Soochong virus (SOOV SOO-1), Dobrava/Belgrade virus (DOB/BGDV Greece), Seoul virus (SEOV 80-39), Tula virus (TULV M5302v), Puumala virus (PUUV Sotkamo), Prospect Hill virus (PHV PH-1), Sin Nombre virus (SNV NMH10) and Andes virus (ANDV Chile9717869). The numbers at each node are posterior node probabilities based on 150,000 trees. The scale bar indicates nucleotide substitutions per site.