Figure 2.
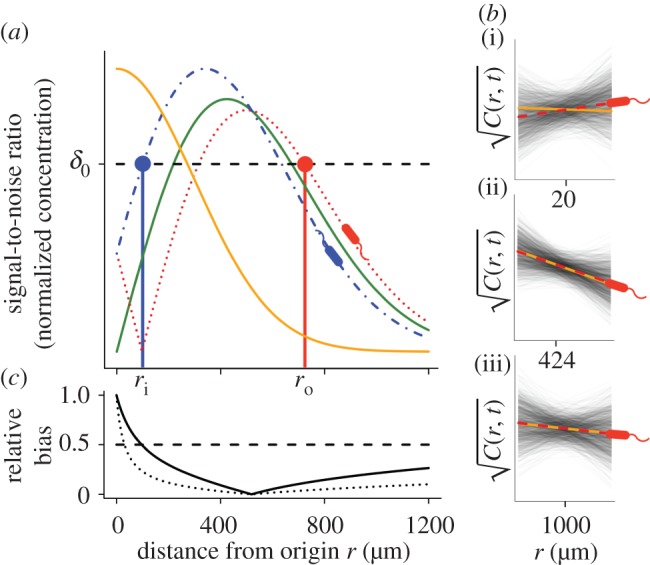
Gradient estimation in a dynamic environment. (a) Solid orange curve shows the true concentration profile at t = t0. Solid green unimodal curve shows the SNR of 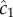 a cell would experience if this concentration profile were static. Dotted red curve shows the SNR for a cell swimming directly towards the origin of the pulse. Dash-dot blue curve shows the SNR for a cell swimming directly away from the origin of the pulse. Concentration and SNR normalized to a maximum value of one. (b) Square root of concentration
a cell would experience if this concentration profile were static. Dotted red curve shows the SNR for a cell swimming directly towards the origin of the pulse. Dash-dot blue curve shows the SNR for a cell swimming directly away from the origin of the pulse. Concentration and SNR normalized to a maximum value of one. (b) Square root of concentration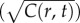 at t = t0 (solid orange line) and individual estimates of this concentration
at t = t0 (solid orange line) and individual estimates of this concentration 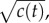 semi-transparent grey; mean of estimates shown by dashed red line) made by a cell swimming towards the pulse origin. Each individual estimate is computed by calculating
semi-transparent grey; mean of estimates shown by dashed red line) made by a cell swimming towards the pulse origin. Each individual estimate is computed by calculating 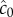 and
and 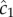 (see the electronic supplementary material for equations) from a time series of random Poisson molecule arrivals [30] with an arrival rate given by the true instantaneous concentration at the bacterium's position C(x, t). (c) Relative bias of concentration slope estimate
(see the electronic supplementary material for equations) from a time series of random Poisson molecule arrivals [30] with an arrival rate given by the true instantaneous concentration at the bacterium's position C(x, t). (c) Relative bias of concentration slope estimate 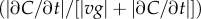 measured by slow (solid curve; v = 30 µm s−1) and fast swimming cells (dotted curve; v = 96 µm s−1). In all panels, the concentration is governed by equation (2.3) with N = 3, M = 1011 molecules, v = 30 µm s−1, a = 1 µm, T = 0.1 s, t0 = 45 s and δ0 = 1. Pulse sizes in all figures correspond roughly to the quantity of free amino acids released from a lysed phytoplankton cell of approximately 10 µm in diameter [5]. (Online version in colour.)
measured by slow (solid curve; v = 30 µm s−1) and fast swimming cells (dotted curve; v = 96 µm s−1). In all panels, the concentration is governed by equation (2.3) with N = 3, M = 1011 molecules, v = 30 µm s−1, a = 1 µm, T = 0.1 s, t0 = 45 s and δ0 = 1. Pulse sizes in all figures correspond roughly to the quantity of free amino acids released from a lysed phytoplankton cell of approximately 10 µm in diameter [5]. (Online version in colour.)
