Abstract
The PTTG1-Binding Factor (PBF/PTTG1IP) has an emerging repertoire of roles, especially in thyroid biology, and functions as a proto-oncogene. High PBF expression is independently associated with poor prognosis and lower disease-specific survival in human thyroid cancer. However, the precise role of PBF in thyroid tumorigenesis is unclear. Here, we present extensive evidence demonstrating that PBF is a novel regulator of p53, a tumor suppressor protein with a key role in maintaining genetic stability, which is infrequently mutated in differentiated thyroid cancer. By coimmunoprecipitation and proximity ligation assays, we show that PBF binds specifically to p53 in thyroid cells, and significantly represses transactivation of responsive promoters. Further, we identify that PBF decreases p53 stability by enhancing ubiquitination, which appears dependent on the E3 ligase activity of Mdm2. Impaired p53 function was evident in a transgenic mouse model with thyroid-specific PBF over-expression (PBF-Tg), which had significantly increased genetic instability as indicated by FISSR-PCR analysis. Consistent with this, ~40% of all DNA repair genes examined were repressed in PBF-Tg primary cultures, including genes with critical roles in maintaining genomic integrity such as Mgmt, Rad51 and Xrcc3. Our data also revealed that PBF induction resulted in upregulation of the E2 enzyme Rad6 in murine thyrocytes, and was associated with Rad6 expression in human thyroid tumors. Overall, this work provides novel insights into the role of the proto-oncogene PBF as a negative regulator of p53 function in thyroid tumorigenesis, where PBF is generally over-expressed and p53 mutations are rare compared to other tumor types.
Thyroid cancer is the fastest increasing cancer in both men and women, with incidence rates rising by 6.6% per year from 1997 to 2009 (1). Genetic alterations that govern cancer initiation and progression have been identified in around 75% of cases, which typically involve effectors of the MAPK and PI3K pathways (2). Despite effective first line treatments to ablate abnormal thyroid tissue, recurrence occurs in approximately 8 to 30% of patients (3, 4), and those genetic alterations driving tumor recurrence remain obscure. Recent progress has been made in associating the BRAF V600E mutation with increased cancer-related mortality among patients with papillary thyroid cancer (5). However, the key molecular mechanisms underlying thyroid carcinogenesis still need to be better defined to enable development of more effective and targeted therapies.
Also known as PTTG1IP, The Pituitary Tumor Transforming Gene 1 binding factor (PBF) is a ubiquitously expressed proto-oncogene that was first identified through its ability to bind the human securin PTTG1 (6). Previously, we showed that subcutaneous expression of PBF induced tumors in athymic nude mice, and that PBF expression was higher in differentiated thyroid carcinomas than in normal thyroid (7). Separate studies have now identified a role for PBF in regulating the sodium iodide symporter (NIS) and the monocarboxylate transporter 8 (MCT8) in thyroid cells (8–10). We also recently demonstrated that thyroid-targeted expression of PBF resulted in the repression of NIS function in vivo, and induced hyperplastic growth and macrofollicular thyroid lesions. In contrast to our previous subcutaneous study, however, thyroid tumor induction was absent in the transgenic mouse model (11). Collectively, these observations indicate that PBF has an emerging repertoire of roles, especially in thyroid biology, and likely functions as a proto-oncogene. Recently the significance of PBF as a prognostic biomarker was also demonstrated in a study of 153 patients with papillary thyroid cancer (12). High PBF expression was significantly correlated with locoregional recurrence, distant metastasis at diagnosis and disease-specific mortality in patients. The precise role of PBF in thyroid tumorigenesis however has not been established.
A critical event in the pathogenesis of most, if not all, human cancers is the disruption of p53 function, a key regulator in maintaining genetic stability (13) and mediating crucial responses to a range of cellular stresses including irradiation-induced DNA damage. For many cancers inactivation of the TP53 gene by somatic mutation typically occurs late in tumor progression, and is often correlated with cancer aggressiveness and poor survival (14, 15). For instance, p53 mutations have been identified in 55% of anaplastic thyroid carcinomas, which have a very poor prognosis due to their aggressive behavior and resistance to cancer treatments (16). This is in stark contrast to well-differentiated thyroid malignancies where the prevalence of p53 mutations is much lower at approximately 11% (13, 17). There is increasing evidence however that p53 inactivation is also caused by physical interactions of p53 with inhibitory proteins which are commonly overexpressed in cancer (18, 19). For instance, the E3 ubiquitin ligase Mdm2/Hdm2 (20, 21) and high mobility group factor A (HMGA) (17, 22) have both been implicated in the pathogenesis of papillary thyroid carcinomas due to their ability to bind and inhibit p53 function (23). It is unclear whether disruption of p53 function by such proteins also contributes towards the increased sensitivity of thyroid glands to radiation-induced oncogenesis (24).
Despite the significant progress made in characterizing proteins that interact with p53, regulation of p53 is highly complex and involves a myriad of different pathways (25, 26). For instance, PTTG1 has been shown to block binding of p53 to DNA thereby inhibiting its ability to induce apoptosis (27), and to cause p53 stimulation and apoptosis following overexpression in MCF-7 cells (28). A more detailed understanding is thus required of the molecular events that occur following inhibition of p53 function which help tumor cells escape the tight regulation of key processes governing survival and proliferation.
Here, we provide extensive evidence that PBF is a novel regulator of p53 function in thyroid cells, and propose that PBF’s mechanism of action in thyroid tumorigenesis is via dysregulation of p53 activity. We have further characterized target DNA repair genes downstream of p53 that are affected by PBF overexpression and have critical roles in maintaining genomic integrity. This study therefore provides a novel insight into understanding the role of PBF in the pathogenesis of thyroid cancer.
Materials and Methods
Cell culture
Primary murine thyrocyte cultures were performed as described (11). Human thyroid papillary carcinoma TPC1 and anaplastic thyroid carcinoma SW1736 cells were kindly provided by Dr Rebecca Schweppe (Division of Endocrinology, University of Colorado Denver), and thyroid papillary carcinoma K1 cells were obtained from the Health Protection Agency Culture Collections. The human nonsmall-cell lung cancer H1299 cells were purchased from American Type Culture Collection (ATCC). All cell lines were kept at low passage number and cultured in RPMI 1640 (Invitrogen) supplemented with 10% FCS. Both TPC1 and K1 papillary carcinoma cells express WT p53 (29, 30), whereas H1299 and SW1736 cells are both p53-null due to either a homozygous partial deletion of the TP53 gene or marked down-regulation of p53 mRNA (31). All cell lines used were confirmed as genuine by STR analysis (DNA Diagnostics Centre, London, UK) following comparison with reported profiles [Supplemental Figure 1; (32)]. Primary thyrocytes were cultured in modified Ham’s F12 media supplemented with 300 mU/L thyrotrophin (Sigma), 100μg/L insulin (Sigma) and 5% FCS. Serum was omitted after 72 hours of culture and experiments performed between 7–11 days of culture.
Animals
Wild-type FVB/N (WT) and transgenic PBF-Tg mice (11) were bred at the University of Birmingham and all experiments performed in accordance with UK Home Office regulations. Thyroids were dissected from 6 week old male PBF-Tg and WT mice for all genetic instability, focused RT2 Profiler PCR array and real-time PCR validation experiments. Expression of thyroidal PBF and Rad6 was examined by immunohistochemistry in aged male and female PBF-Tg mice with a mean age of 15 months (461 ± 44 days; n = 9). Similar trends in protein expression were observed for male and female PBF-Tg mice compared to age and sex-matched WT controls.
Human thyroid samples
Matched tumor and normal tissue specimens were obtained from 11 patients undergoing surgery for thyroid cancer at the University Hospital Birmingham NHS Trust, UK. Normal specimens were taken from the contralateral lobe at the time of surgery and were shown to be noncancerous upon histological examination. All specimens harvested at the time of resection were collected with appropriate local ethical committee approval and informed patient consent.
Transfections
Plasmid DNA and siRNA transfections were performed with Fugene6 (Roche) and Lipofectamine-2000 (Invitrogen) according to manufacturer’s instructions. Cells were transfected using pooled PBF-specific siRNA (#14399 and #147350), pooled Rad6-specific siRNA (#4390824 and #4390825) or negative control siRNA (AM4635) at a final concentration of 100 nM (Ambion). For plasmid DNA experiments cells were transfected with pcDNA3 containing full-length wild-type PBF cDNA with a hemagglutinin (HA)-tag, the Rad6 expression vector pCMV6-Ube2a (Cambridge Biosciences, UK) or empty pcDNA3 (Invitrogen) as vector only (VO) unless otherwise stated, using conditions previously described (9).
Western blot
Western blot analyses were performed as described previously (9, 33). Blots were probed with specific antibodies against PBF (9, 33), 1:200; HA (Covance Research Products), 1:2000; p53(D0–1) (Santa Cruz Biotechnology), 1:1000 and Rad6 (Abcam, #ab31917), 1:1000. Antigen-antibody complexes were detected using the ECL Plus chemiluminescent detection system (Amersham Biosciences). Actin expression was determined using mouse monoclonal anti-β actin antibody clone AC-15 (Sigma-Aldrich) at 1:10,000. Protein quantification was performed on cell lysates using the Bradford assay. To quantify detected bands by densitometry, blots were scanned into Photoshop (Adobe Systems) keeping all scanning parameters the same and analyzed using ImageJ software (34).
Binding assays
WT p53 and deletion mutants were cloned into pGEX4T-1 for bacterial expression. L-α-[35S]-methionine-labeled PBF was expressed in vitro using a TNT T7 Coupled Reticulocyte Lysate System according to the manufacturer’s guidelines (Promega). In vitro glutathione-S-transferase (GST) pull-down assays using [35S]-PBF and GST-p53 proteins were performed using established protocols (35). GST pull-down and coimmunoprecipitation (co-IP) assays were performed as described previously (9). The Duolink in situ proximity ligation assay (PLA) was performed according to manufacturer’s instructions (Olink Bioscience). In our experiments thyroid cells were seeded onto coverslips and transfected with expression vectors for p53 (pcDNA3-p53) and HA-tagged PBF (pcDNA3-PBF) 24 hours later prior to the PLA assay.
p53 stability assays
In p53 half-life experiments cells were incubated in 100 μM anisomycin for 2 hours prior to cell lysate extraction using standard protocols. Nutlin-3 (Sigma-Aldrich) was added to cells at 50 μM concentration for 6 hours prior to assessment of p53 stability. In p53 ubiquitination experiments, cells were incubated in 20μM MG132 for 2 hours prior to cell lysate extraction.
qRT-PCR and sequencing
Total RNA was extracted using the RNeasy Micro Kit (Qiagen) and reverse transcribed using the Reverse Transcription System (Promega). Expression of specific mRNAs was determined using 7500 Real-time PCR system (Applied Biosystems). Gene expression of total RNA extracted from primary thyrocytes was analyzed using the DNA damage signaling pathway-focused RT2 Profiler PCR Array (SABiosciences) according to manufacturer’s instructions. cDNA was sequenced to determine p53 mutational status of human thyroid tumors essentially as described (36). Primer sequences are listed in Supplementa1 Figure 8.
Irradiation and genetic instability assays
DNA damage was induced by Caesium 137 irradiation using an irradiator IBL 437C type H unit (CIS Bio international, Gif Sur Yvette). FISSR-PCR amplifications were performed essentially as described previously (37) using a 5′ FAM-labeled primer (CA)8RG with 5 ng of genomic DNA. PCR products were electrophoresed on an ABI3730 capillary sequencer (Applied Biosystems) and data analyzed using Peak Scanner v1.0 software. Five replicate experiments were performed to verify the reproducibility of the assay. The degree of genetic instability was determined according to Basik et al (38) to generate the genetic instability index (GI), which represents the standard measure of genetic instability with ISSR-PCR analysis.
Reporter gene assays
The p53 reporter plasmids phdm2-Luc and p21-Luc have been described previously (39), and contain fragments of either the hdm2 or p21 promoter linked to the firefly reporter gene. H1299 cells were transfected with 400 ng pCIneo-PBF (PBF), 5 ng pcDNA3-p53 (p53), and 150 ng of the indicated p53 reporter plasmid. The empty plasmid pCIneo without the PBF cDNA was used as vector only (VO) as indicated. The Renilla luciferase control plasmid pRL (Promega) was used (20 ng per transfection) as an internal control to normalize firefly luciferase expression. Cells were harvested in Passive Lysis Buffer and the Dual Luciferase Reporter Assay System (Promega) used to measure luciferase activity. Data was normalized to Renilla activity.
Immunohistochemistry and analysis of thyroid morphology
Thyroid glands were removed from mice using a dissecting microscope. Tissue was fixed in 10% formal saline for at least 24 hours prior to being paraffin-embedded and cut into 5 μm sections according to standard protocols. Formalin-fixed paraffin embedded sections of mouse thyroid tissue was immunostained using an avidin-biotin peroxidase technique (Vectastain Elite, Vector laboratories, Burlingame, CA). Immunostaining was performed with specific antibodies against HA, (Covance Research Products), 1:200; and Rad6, (Abcam, #ab84395), 1:250 using protocols as described previously (11). For negative controls the primary antibody was replaced by 10% normal goat serum. Antigen retrieval was performed to enhance immunostaining of murine thyroids by heating tissue sections in boiling 10 mM citrate buffer (pH 6.0) for 10 minutes on a low setting in a domestic microwave (Panasonic, NN6453, 800W). Hematoxylin and eosin (H&E) and immunostained thyroid tissue specimens were viewed under a light microscope (Zeiss) and images captured using Axiovision software (Version 4). A standard 100 μm scale bar (Axiovision) was used to convert pixels to μm.
Cell Survival and Apoptosis Assays
The cellular viability of TPC1 and SW1736 cultures was determined using the CellTiter 96 AQueous One Solution Cell Proliferation (MTS) assay (Promega) according to the manufacturer’s instructions. MTS absorbance readings were determined using a Victor3 plate reader (Perkin Elmer) after incubation for 1 hour at 37°C in a humified chamber containing 5% CO2. Relative levels of apoptosis were determined by measuring caspase 3 and 7 enzyme activity using the Caspase Glo 3/7 Apoptosis Kit (Promega) and assays performed according to the manufacturer’s instructions.
Statistical analysis
Data are displayed as mean ± SE. Normally distributed data were analyzed using a two-tailed Student’s t test, unless otherwise indicated. A P-value < 0.05 was considered to be statistically significant.
Results
PBF binds to p53
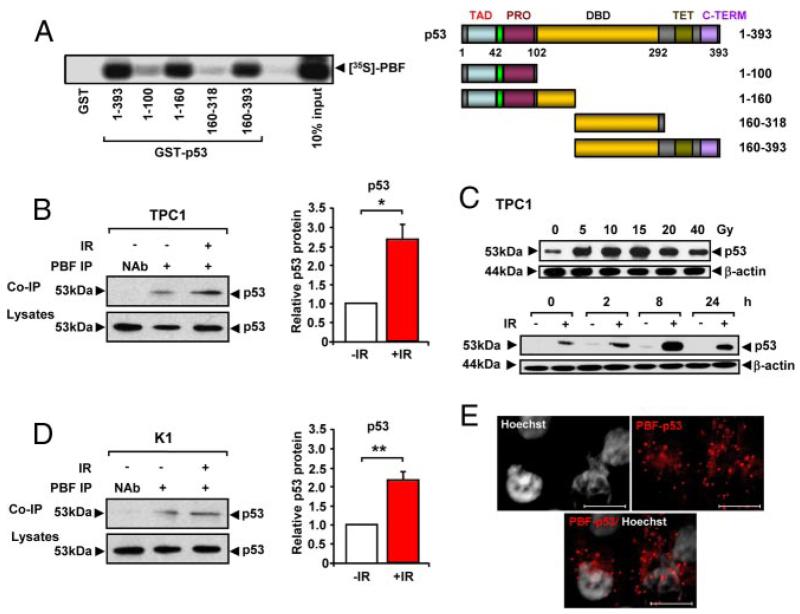
To investigate a role for PBF in thyroid tumorigenesis we examined whether PBF interacts with p53, a protein critical in suppressing human cancers. Initial GST pull-down assays demonstrated that L-α-[35S]-methionine-labeled PBF binds to the p53 protein (Figure 1A). Successive deletion mutants of GST-p53 altered the stringency of PBF binding. For instance, PBF did not bind efficiently to the N-terminus region of p53 located between amino acids 1 to 100. Instead the strongest binding sites for PBF appeared to be located within residues 100–160 and 318–393 of p53 (Figure 1A). Endogenous coimmunoprecipitation (co-IP) assays in papillary thyroid cancer TPC1 and K1 cells confirmed that p53 specifically associates with PBF (Figure 1B).
Figure 1.
Interaction between PBF and p53. A, Binding of [35S]-PBF to GST-p53 (1–393) and GST-p53 deletion mutants as indicated vs a GST-only control. Schematic of domain structure of p53 and deletion mutants showing a transcriptional activation domain (TAD), proline rich domain (PRO), DNA-binding domain (DBD), tetramerization domain (TET) and C-terminal (C-TERM). B, Co-IP of p53 with PBF in TPC1 cells either untreated (−IR) or irradiated with 15 Gy for 8h (+IR). NAb=no antibody control. Graph shows quantified levels of p53 ± SE from 3 independent experiments. C, Western blot analysis of p53 in TPC1 cells irradiated with 0 to 40 Gy dose as indicated for 8 hours (upper), or irradiated (+) with 15 Gy dose and p53 protein levels monitored at 0, 2, 8 or 24 hours post-treatment compared to untreated (−) controls (lower). D, Co-IP of p53 with PBF in K1 cells either untreated (−IR) or irradiated with 15 Gy for 8h (+IR). NAb=no antibody control. Graph shows quantified levels of p53 ± SE from 3 independent experiments. E, PLA assay to demonstrate specific PBF and p53 interaction (red spots) in TPC1 cells transiently transfected for 24 hours with plasmid expression vectors for p53 and HA-tagged PBF. Scale bars: 10 μM. *, P < .05; **, P < .01.
p53 is maintained at low cellular levels but stabilized by irradiation-induced DNA damage. We therefore determined the radiation dose and timing required to yield an optimal p53 response in both TPC1 and K1 cells (Figure 1C and Supplemental Figure 2) and examined the subsequent interaction between p53 and PBF. Irradiation treatment using these optimal conditions (ie, 15 Gy, 8 hours) led to an increased quantity of coimmunoprecipitated p53 with PBF in both TPC1 (P < .05; ~2.7-fold; Figure 1B) and K1 (P < .01; ~2.2-fold; Figure 1D) cells compared to controls. Proximity ligation assays [PLA; (40)] also demonstrated the presence of red spots of specific p53 and PBF interaction after transient transfection in both TPC1 (Figure 1E and Supplemental Figure 3 A and B) and K1 cells (Supplemental Figure 3C).
PBF increases turnover and ubiquitination of p53
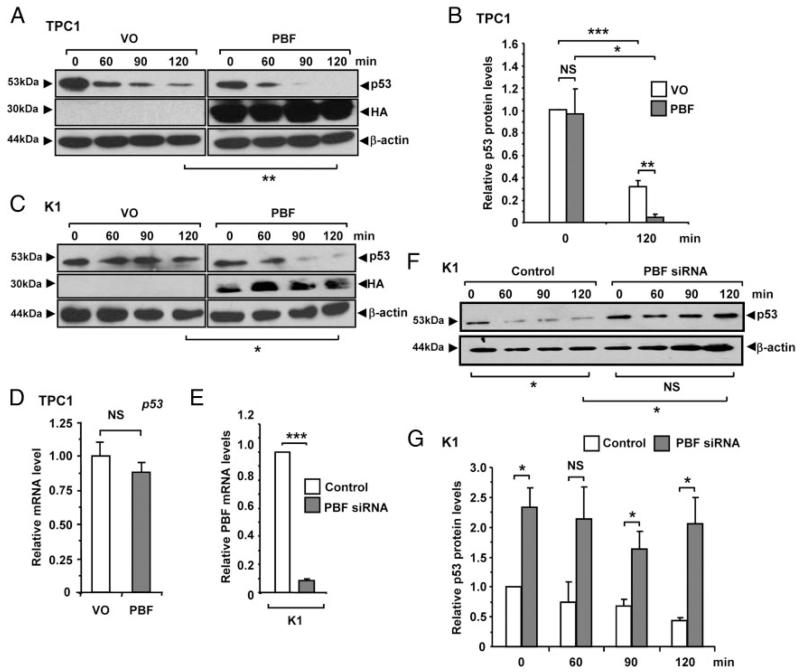
p53 is an intrinsically unstable protein which is subject to rapid degradation, and is rarely mutated in differentiated thyroid cancer. To further investigate the relationship between PBF and p53 in thyroid cells, we examined whether interaction with PBF resulted in altered p53 stability. Half-life studies using anisomycin to block de novo protein synthesis showed that overexpression of PBF significantly increased turnover of p53 protein in TPC1 (P < .01; Figure 2, A and B) and K1 (P < .05; Figure 2C) cells compared to vector only (VO) controls after 120 minutes. Importantly, control experiments showed that p53 mRNA levels did not change significantly in TPC1 cells following overexpression of PBF compared to VO (Figure 2D). To determine if the reverse relationship held true, we depleted PBF (Figure 2E) and observed greater p53 stability, with > 4-fold higher p53 levels (P < .05) in K1 cells compared to negative controls after 120 minutes, and no significant turnover over the timecourse (Figure 2, F and G). Increased p53 stability was also evident in PBF-depleted TPC1 cells compared to controls at 120 minutes postanisomycin treatment (data not shown).
Figure 2.
PBF decreases p53 intracellular stability. Representative Western blot analysis of p53 in (A) TPC1 and (C) K1 cells transfected with either vector only (VO) or PBF and then lysed at indicated times post-treatment with 100 μM anisomycin. Detection of HA epitope was used to monitor transfection. Mean p53 protein levels relative to β-actin in TPC1 cells are shown in (B) from 3 independent experiments. D, Relative mRNA levels of p53 in TPC1 cells transfected with either VO or PBF. E, Relative mRNA levels of PBF in K1 cells transfected with either PBF-specific or control siRNA for 72 hours at a concentration of 100 nM. F, Representative Western blot analysis of p53 in K1 cells transfected with either PBF-specific or control siRNA and then lysed at indicated times post-treatment with 100 μM anisomycin. G, Quantification of p53 protein levels relative to β-actin from p53 half-life experiments in K1 cells transfected with either PBF-specific or control siRNA. Data presented as mean p53 levels ± SE from 3 independent experiments. *, P < .05; ***, P < .001; NS = not significant.
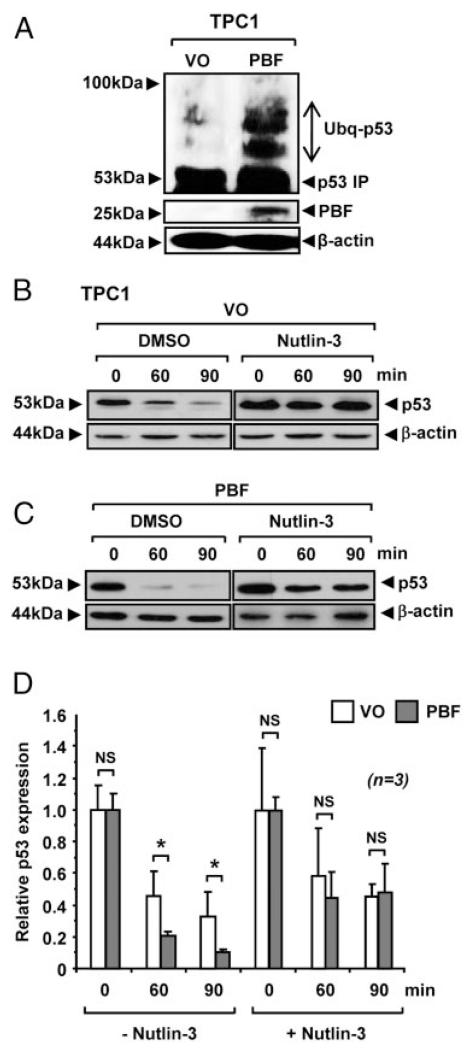
Given that modulation of PBF expression was associated with altered p53 stability, we examined p53 ubiquitination in cells treated with the proteasome inhibitor MG132. A significant increase in the level of high mwt p53 conjugates was present in PBF-transfected TPC1 cells treated with MG132 (Figure 3,A), which is consistent with the accumulation of ubiquitinated p53. Mdm2 has been identified as the major E3 ubiquitin ligase of p53, binding and targeting it for proteasome-mediated degradation. We therefore investigated whether the increased turnover of p53 by PBF in thyroid cells was dependent on the E3 ligase activity of Mdm2. The addition of the inhibitor nutlin-3 to block binding of Mdm2 to p53 led to a significant increase in p53 stability in VO- (Figure 3B) and PBF-transfected TPC1 cells (Figure 3C). Quantification of p53 protein showed that equivalent levels of p53 were present in VO- and PBF-transfected TPC1 cells at both 60 and 90 minutes post-treatment with nutlin-3 (P = NS; n = 3; Figure 3D). This was in contrast to a significant decrease in p53 stability in the absence of nutlin-3 in PBF-transfected cells at the same time points (P < .05; Figure 3D). Therefore, the ability of PBF to diminish p53 stability in thyroid cells appears to be Mdm2-dependent with no evidence of any additive or synergistic interactions.
Figure 3.
Effect of PBF on p53 ubiquitination. A, Detection of high mwt p53 conjugates by Western blot analysis in TPC1 cells transfected with either VO or PBF and then treated with 10 μM MG132. B-C, Western blot analysis of p53 in TPC1 cells transfected with either VO or PBF and then incubated with 50 μM nutlin-3 prior to 100 μM anisomycin treatment. DMSO was used as vehicle. D, Mean p53 protein levels relative to β-actin quantified from 3 independent experiments are shown. Data presented as mean ± SE. *, P < .05; NS = not significant.
PBF inhibits p53 activity and alters sensitivity of thyroid cells to irradiation
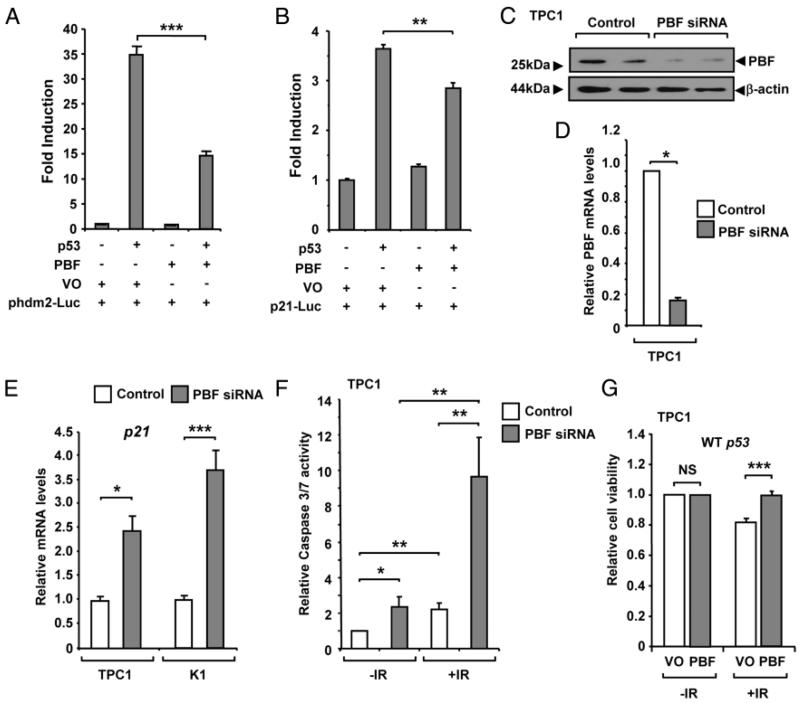
Following our observations that PBF binds and regulates p53 stability, we next analyzed whether PBF altered the transactivation activity of p53 in transient reporter assays in H1299 cells, which are p53-null. When coex-pressed, PBF significantly repressed p53-mediated hdm2 promoter activity by ~60% (P < .001; Figure 4A), as well as p21 promoter activity (~25%; P < .01; Figure 4B). We next depleted PBF (Figure 4, C and D) and observed a significant increase in the mRNA expression of p21, a well-characterized p53 responsive gene, in both TPC1 (2.4-fold; P < .05) and K1 (3.7-fold; P < .001) papillary thyroid cells (Figure 4E). Examination of apoptotic markers also indicated a corresponding increase in caspase-3/7 activity in PBF-depleted TPC1 (2.4-fold; P < .05; Figure 4F) and K1 cells (1.45-fold; P < .05; Supplemental Figure 4A).
Figure 4.
PBF inhibits p53 transcriptional activity. H1299 cells were transfected with p53 luciferase reporter plasmids for either (A) hdm2 (phdm2-Luc) or (B) p21 (p21-Luc), as well as p53 and PBF expression vectors, or vector only (VO) as indicated. Luciferase activity was measured 24 hours post-transfection. C-D, TPC1 cells were transfected with either PBF-specific or control siRNA at a final concentration of 100 nM. PBF expression was assessed by Western blotting and qRT-PCR analysis as shown. E, Relative mRNA levels of p21 mRNA in TPC1 and K1 cells transfected with either PBF-specific or control siRNA for 48 hours at a concentration of 100 nM. F, Analysis of caspase-3/7 activity in TPC1 cells transfected with either PBF-specific or control siRNA for 48 hours and then irradiated with a 15 Gy dose (+IR) or untreated (−IR). Normalized mean caspase-3/7 values ± SE are shown from 4 independent experiments. For each experiment caspase-3/7 activity was determined from n = 5 per condition at 24 hours postirradiation. G, TPC1 cells were transfected with either VO or PBF for 24 hours and either untreated (−IR) or irradiated (+IR) with a 15 Gy dose. Cells were then replated and viability measured after 24 hours. Data presented as mean ± SE from four independent experiments. *, P < .05; **, P < .01; ***, P < .001; NS = not significant.
To further investigate the physiological relevance of our finding that PBF alters p53 stability and activity, we next examined the influence of manipulating PBF expression on apoptosis and cell survival in response to irradiation. Interestingly, depletion of PBF in TPC1 cells appeared to enhance their sensitivity to irradiation with a greater fold-change in caspase-3/7 activity (4.35-fold; P < .01; Figure 4F) and a significant decrease in cell viability (P < .05; Supplemental Figure 4B). Furthermore, in the absence of PBF, TPC1 cell survival was reduced by ~20% in response to irradiation (Figure 4G), whereas cells transfected with PBF demonstrated no decrease in cell survival (P < .001). Importantly, there was no difference in the cell viability of irradiated p53-null SW1736 thyroid cells transfected with either VO or PBF (P = NS; Supplemental Figure 4C), suggesting that the effects of PBF on cell survival may be p53 dependent. Together these results support the notion that the functional consequences of PBF dysregulation are via regulation of p53 activity.
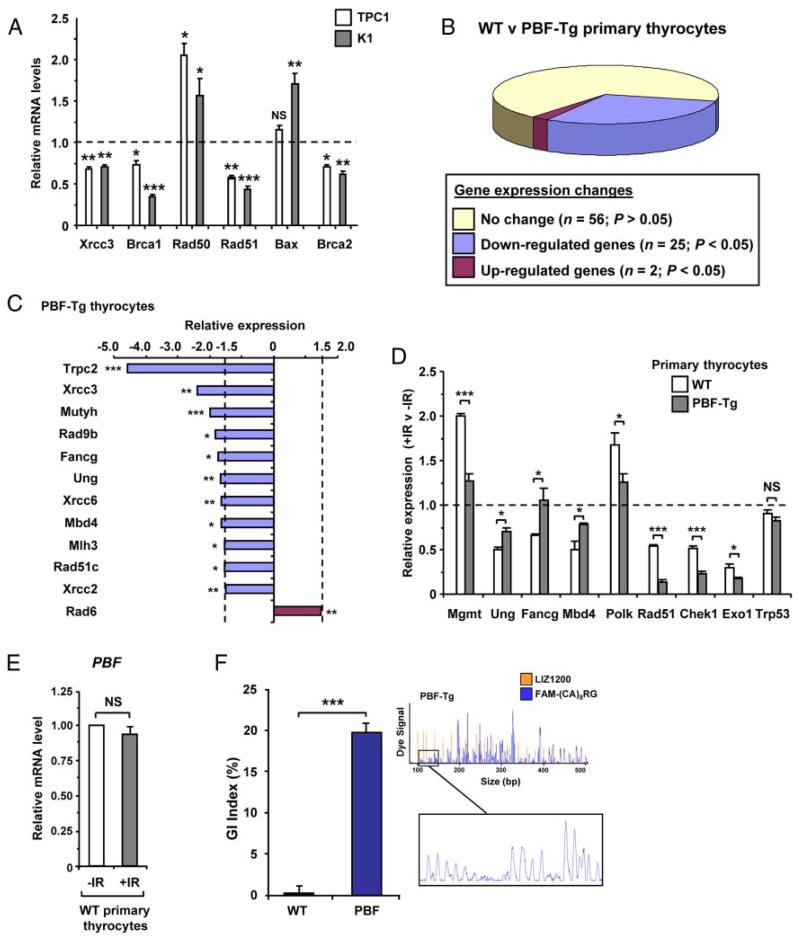
PBF dysregulates DNA repair genes and promotes genetic instability
We next examined whether PBF depletion in thyroid cell lines also caused dysregulation of other p53-responsive genes. Figure 5A shows that significant mRNA changes were indeed observed in PBF-depleted TPC1 and K1 cells for a number of other genes including DNA repair genes such as Rad50, Rad51, Brca1 and Brca2. To understand the effect of PBF dysregulation in a more physiologically relevant thyroid-disease model, we next examined the transcriptional profile of a panel of 83 p53-regulated genes in murine primary PBF-Tg thyrocytes compared to wild-type (WT) thyrocytes (Figure 5B). A total of 27 genes showed significant expression changes (P < .05), suggesting wide-ranging dysregulation in response to raised PBF. Of these, 12 genes showed altered mRNA expression > 1.5-fold (Figure 5C), including several genes known to maintain genomic integrity, such as Xrcc3 (0.42 ± 0.09; P = .003), Fancg (0.58 ± 0.1; P = .02) and Rad51c (0.65 ± 0.09; P = .02).
Figure 5.
PBF dysregulates DNA repair gene expression and promotes genetic instability. A, Relative mRNA expression of indicated genes in TPC1 and K1 cells transfected with either PBF-specific or control siRNA for 48 hours at a concentration of 100 nM. B, Pie chart summarizes expression changes of DNA repair genes between PBF-Tg and WT thyrocytes (n = 3 arrays). C, Relative mRNA expression levels of 12 genes (≥ 1.5-fold; P < .05) in PBF-Tg thyrocytes compared to WT. D, Relative fold changes in mRNA expression of indicated 9 genes following irradiation of either WT or PBF-Tg thyrocytes compared to nonirradiated controls as indicated. Data presented as mean ± SE from at least 3 independent experiments. E, Relative mRNA levels of PBF in wild-type (WT) primary thyrocytes either untreated (−IR) or irradiated with a 15 Gy dose (+IR). F, Quantification of genetic instability in murine PBF-Tg thyrocytes compared to WT (n = 5). Data presented as mean GI index ± SE. A representative FISSR-PCR trace amplified from PBF-Tg thyrocyte gDNA is shown plotted against a LIZ1200 size standard. *, P < .05; **, P < .01; ***, P < .001; NS = not significant.
Primary thyrocyte cultures from PBF-Tg mice were next irradiated in order to identify additional p53-responsive candidate genes that might be disrupted by elevated endogenous PBF in nontransformed cells. A total of ten genes, including Rad51, Chek1 and Rad6, showed significant changes (> 1.5-fold; P < .05) in their mRNA levels following irradiation of PBF-Tg thyrocytes compared to WT (Supplemental Figure 5). Normalization of data to adjust for PBF effects on gene expression identified 8 genes most significantly dysregulated by elevated PBF following irradiation (Figure 5D). For example, irradiation-induced expression of the p53-responsive genes Mgmt (2.0 ± 0.03-fold) and Polk (1.7 ± 0.08-fold) was significantly suppressed in PBF-Tg thyrocytes compared to WT (P = .0007 and P = .05 respectively; Figure 5D). In contrast, the degree of irradiation-induced inhibition for several genes, including Ung (P = .02) Fancg (P = .05) and Mbd4 (P = .04), was abrogated in PBF-Tg thyrocytes following irradiation. Validation experiments using individual qPCR assays also confirmed significant differences in expression of Mgmt (P = .009), Rad51 (P = .0002) and Chek1 (P = .002) in PBF-Tg thyrocytes postirradiation (Supplemental Figure 6). Furthermore, irradiation did not cause any transcriptional changes in either TP53 (Figure 5D) or PBF (Figure 5E).
To investigate whether the ability of PBF to abrogate p53 activity and downstream DNA repair genes might also influence genetic stability in vivo, we next used fluorescent inter simple sequence repeat (FISSR) PCR (37) to analyze genetic instability in thyroid glands dissected from PBF-Tg and WT mice. In comparison to age and sex-matched WT mice [arbitrarily assigned a genetic instability (GI) of 0%], the thyroids of 6-week old PBF-Tg mice demonstrated significantly disrupted genomes, with a mean GI index of 19.8 ± 1.8% (P < .001; Figure 5F). Altogether these results identify a panel of p53-responsive DNA repair genes that are dysregulated by elevated PBF levels in primary thyrocytes, and are associated with genetic changes underlying the increased GI index observed in PBF-Tg thyroids. Among these, the expression of Rad6, also known as the ubiquitin-conjugating enzyme E2A (Ube2a), was the sole gene up-regulated > 1.5-fold (P = .008) in PBF-Tg thyrocytes.
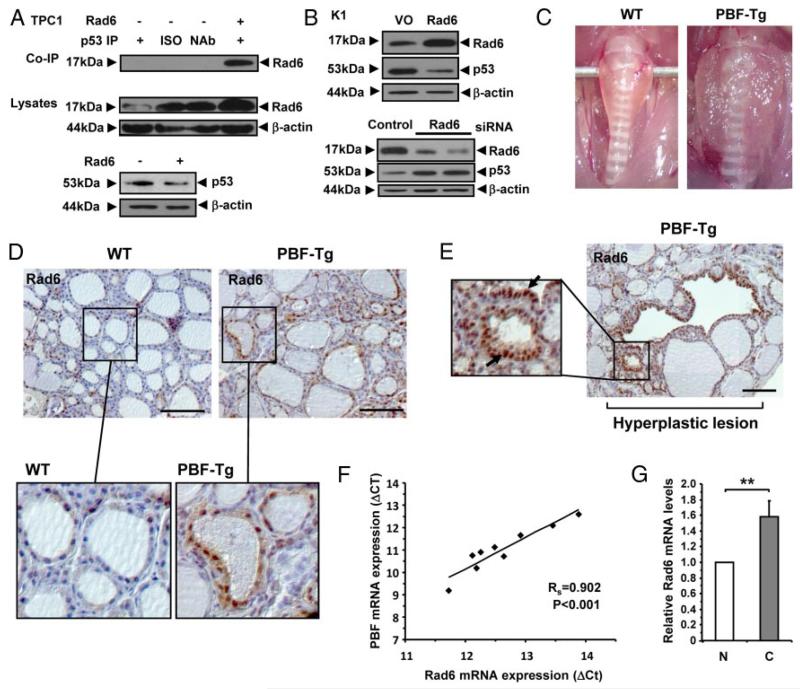
Functional interaction between p53 and Rad6 in thyroid cells
Recently, Rad6 has been shown to form a ternary complex with Mdm2 and p53 that contributes to p53 degradation in HeLa cells (41). We therefore examined whether Rad6 also binds to p53 in thyroid cells. Exogenous coimmunoprecipitation assays confirmed that Rad6 specifically binds to p53 in TPC1 cells (Figure 6A). Importantly, overexpression of Rad6 in both TPC1 (Figure 6A) and K1 (Figure 6B) thyroid cells led to a significant decrease in p53 protein levels. In addition, depletion of Rad6 in K1 cells increased p53 protein levels (Figure 6B).
Figure 6.
Association of Rad6 and PBF in vivo. A, Western blot analysis (upper) of co-IP of Rad6 with p53 in TPC1 cells transfected for 24 hours with either VO (−) or pCMV6-Ube2a (+). ISO = isotype control antibody. NAb = no antibody control. Western blot analysis of Rad6 (middle) and p53 (lower) in cell lysates from transfected TPC1 cells are also shown. B, Western blot analysis of Rad6 and p53 following either overexpression (upper) or depletion (lower) of Rad6 in K1 cells. Control lanes are indicated. C, Representative enlarged thyroids dissected from aged PBF-Tg mice compared to WT. Thyroids were typically ~3–4 fold heavier in PBF-Tg mice. D-E, Representative images of Rad6 staining in normal regions (D) and hyperplastic lesions (E) in WT (n = 6) and PBF-Tg (n = 9) thyroids are shown. Arrows highlight elevated nuclear expression of Rad6 (E). F, Correlation of PBF and Rad6 mRNA expression in human thyroid tumors (n = 11). Statistics analyzed using Spearman rank correlation. G, Graph shows quantification of Rad6 mRNA expression in thyroid tumors relative to normal thyroid. Scale bars: 100 μm. Data presented as mean ± SE. **, P < .01.
Having established that a functional interaction exists between Rad6 and p53 in thyroid cells, we next examined the relationship between PBF and Rad6 in thyroid disease models in vivo. For these experiments we used thyroid glands dissected from PBF-Tg mice, which have an enlarged and hyperplastic phenotype [Figure 6C; (11)]. Immunostaining revealed elevated Rad6 expression in thyroids from 15 month old PBF-Tg mice compared to age-matched WT thyroids (Figure 6D and Supplemental Figure 7A). Previous studies have indicated weak cytoplasmic Rad6 staining in normal cells compared to intense nuclear reactivity in invasive tumor cells (42). Here, Rad6 expression was most prominent in nuclear compartments of follicular epithelial cells in PBF-Tg thyroids; whereas WT cells displayed mostly diffuse cytoplasmic staining (Figure 6D). Importantly, the greatest abundance of Rad6 nuclear immunoreactivity was detected in distinct hyperplastic lesions (Figure 6E), which also colocalized with highest expression of the HA-tagged PBF transgene (Supplemental Figure 7B). In further studies, evaluation of matched normal and tumor human thyroid specimens also revealed a significant positive correlation between PBF and Rad6 mRNA expression (Figure 6F; P < .001; rs = 0.9), with a significant 1.6 ± 0.2-fold induction (P = .008) in Rad6 mRNA levels in thyroid tumors compared to normal tissue (Figure 6G). cDNA sequence analysis showed all human thyroid tumors had WT p53 (Supplemental Figure 8). Taken together, these results suggest a significant association between elevated PBF and Rad6 expression in thyroid disease with WT p53 status. The low mutation rate of the p53 gene in differentiated thyroid cancer may therefore belie functional inactivation of the protein by PBF, which is upregulated in differentiated thyroid cancer, increases p53 turnover, and induces the expression of Rad6, a known regulator of p53 activity.
Discussion
The functional disruption of p53 activity has a critical role in promoting tumorigenesis in many different types of cancer (15). However, the mechanisms governing p53 inactivation remain to be fully defined, especially in tumors such as in differentiated thyroid cancer that have a lower incidence of p53 mutations (16, 17). Our current findings now indicate that the role of the relatively uncharacterised gene PBF in cell transformation most likely reflects its interaction with p53, thus providing a novel insight into the ability of PBF to promote endocrine tumorigenesis.
There has been relatively little reported concerning a possible role for PBF in tumorigenesis, despite identification of the PBF gene in 1998 (43). We previously described PBF overexpression in thyroid (7), pituitary (44) and breast cancers (33). Subsequent functional studies highlighted that PBF was a transforming gene in vitro and induced subcutaneous high-grade malignant tumor formation in athymic nude mice (7). However, the underlying role for PBF in tumorigenic growth in vivo remains unclear, especially as transgenic mice with thyroid-specific PBF expression did not develop thyroid cancer (11).
To therefore gain further insight into the role of PBF in tumorigenesis we investigated the ability of PBF to bind p53 by GST pull-down, coimmunoprecipitation and proximity ligation assays. Our data showed that PBF bound specifically to p53 in vitro and the relative level of p53:PBF coimmunoprecipitates in thyroid cells was enhanced by γ-irradiation. These results most likely reflect the greater abundance of stabilized p53 protein following irradiation as p53 protein levels were, as expected, increased in irradiated TPC1 and K1 cells, in contrast to minimal changes in PBF protein (data not shown).
It is well-documented that proteins such as Mdm2 and USP10 (45) can bind p53 and target it for degradation or promote its intracellular stability. This led us to investigate whether the ability of PBF to interact with p53 might also alter its turnover. Our results indicated that PBF has a role in diminishing p53 stability as PBF overexpression significantly increased p53 turnover in thyroid cancer cells. Further evidence was provided by depleting PBF protein, which increased p53 stability, as well as the enhanced ubiquitination of p53 in PBF overexpressing cells as evidenced by the accumulation of high mwt p53 conjugates. These findings might infer a direct role for PBF in ubiquitinating p53 but the amino acid sequence for PBF does not contain a typical ubiquitin-conjugating (UBC) (46) or E3 ligase domain (47). Additionally, p53 degradation was blocked by the inhibitor nutlin-3 with no synergistic or additive effects in the presence of elevated PBF, thereby indicating that the effects of PBF on p53 ubiquitination may involve Mdm2.
GST pull-down assays using deletion mutants of p53 highlighted two possible regions that might be involved in binding PBF. One of these, located between residues 318 to 393, contains the key lysines ubiquitinated by Mdm2 (48), and which binds the N-terminal domain of Mdm2 (49). We therefore envisage that PBF may modulate the association of p53/Mdm2 with interacting proteins such as HDACs (50), PCAF (51), p300/CBP (51) and Tip60 (52) that are known to alter p53 acetylation and ubiquitination status. The regulation of Mdm2 is however a focal point of numerous regulatory pathways for p53, including both transcription and nontranscriptional targets of p53 (26). For example, the transcription factor YY1 enhances p53 degradation by increasing the binding of Mdm2 to p53 (53). Future investigations will thus need to focus on the precise functional relationship between PBF and p53/Mdm2-interacting partners, as well as the interaction between Mdm2 and p53 itself.
Previous studies have identified that PBF can interact with the proto-oncogene PTTG1 to facilitate its translocation into the nucleus (6). Hence, it is possible that an alternative mechanism exists such that PBF might augment PTTG1’s nuclear function as the human securin, inhibiting mitosis and generating intrachromosomal breaks, as well as increasing the interaction between PTTG1 and p53 (27, 28, 54). Indeed, we have also reported that PTTG1 induces genetic instability in colorectal cells, and that PTTG1 expression correlates with genetic instability in vivo (37). Against this, our current data reveal the novel finding that PBF and p53 bind specifically, particularly in the presence of DNA damage, and that p53 stability was significantly altered by PBF. Furthermore, preliminary co-IP experiments showed that the PBF:p53 interaction was not significantly altered in PTTG1-depleted TPC1 cells (data not shown). These results therefore imply an independence of action for PBF.
Disruption of p53 by interaction with other proteins typically results in loss of activity of p53-responsive genes. Our data of reduced transactivation of Hdm2 and p21 promoters in p53-null H1299 cells transfected with PBF and p53 are therefore in keeping with those in the literature. Indeed, the magnitude of this inhibition was broadly equivalent to proteins such as Polo-like kinase 1 (Plk1) that also physically interact with p53 (55). Importantly, we showed that depletion of PBF in thyroid cells also increased expression of the p53-responsive gene p21, further emphasizing the role of PBF as a negative regulator of p53 activity. Our group recently reported the generation of a PBF-Tg transgenic mouse with targeted human PBF over-expression in the thyroid gland (11). In this study, we were able to show dysregulated expression of approximately 40% of all DNA repair genes examined (32 out of 83) in PBF-Tg primary thyrocytes. It will be important to determine the relative contribution of p53-independent pathways, if any, to the ability of PBF to disrupt expression of these genes. However, expression of many of the genes identified such as Mgmt are known to be modulated by ionizing radiation in a classical WT p53 gene–dependent manner (56), thus the ability of PBF to abrogate these genes provides an invaluable starting point for defining a role for PBF in regulating p53 pathways.
Higher levels of mRNA encoding the E2 enzyme Rad6 in PBF-Tg thyrocytes highlighted a potential route via which PBF might disrupt p53 stability. Subsequent examination of PBF-Tg thyroid glands revealed an increased abundance of nuclear Rad6 protein, a feature of invasive tumor cells (42), which colocalized with high PBF expression, especially at hyperplastic lesions. Recently it was shown that Rad6 plays a critical role in regulating p53 protein levels by forming a ternary complex with Mdm2 and p53 that contributes to p53 degradation in HeLa cells (41). Our results therefore indicate that PBF might also upregulate Rad6 expression in order to decrease p53 stability. The precise mechanism by which PBF induces Rad6 is the subject of further work, although p53 response elements are located in the Rad6 promoter (57). However, we were able to confirm for the first time in thyroid cells that Rad6 coimmunoprecipitates with p53 and overexpression of Rad6 decreases p53 protein levels. In addition, a significant correlation was evident between PBF and Rad6 mRNA expression in human thyroid cancer specimens, further emphasizing the interaction between these two genes in thyroid disease.
Genomic instability is an evolving hallmark of cancer but the molecular basis is poorly defined (58). In our study elevated genetic instability was present in PBF-Tg mouse thyroids and associated with extensive repression of DNA repair genes. Of particular importance, the disruption of several genes highlighted in this study, including the Rad51 family (ie, Rad51, Xrcc3, Xrcc2 and Rad51c), have been associated with an increased risk of thyroid cancer (59). Furthermore, inactivation of Mgmt has been suggested to cause somatic mutations in RAS (60), a gene commonly mutated in thyroid neoplasms. Our results in this study would therefore suggest a novel mechanism for tumorigenesis such that PBF overexpression may disrupt DNA repair enzymes with critical roles in the pathogenesis of cancer, as well as contributing to the overall genetic instability typically associated with tumor progression (61).
We previously showed that thyroid cancers displaying recurrence demonstrate particularly high PBF levels (7). Similarly, a recent study showed that high PBF expression was significantly correlated with locoregional recurrence and distant metastases at diagnosis in patients with papillary thyroid cancer (12). These findings support a larger-scale clinical evaluation to determine whether PBF expression in thyroid cancer correlates with genetic instability and somatic mutations typically associated with aggressive disease and increased mortality such as BRAF V600E (5)
In summary, we describe PBF as a novel interacting partner of p53, which alters p53 stability and transactivation capabilities. In defining a role for PBF, we demonstrate that it interferes with DNA repair pathways and induces genetic instability in thyroid cells in vivo. This work therefore provides an important insight into the role of the proto-oncogene PBF as a negative regulator of p53 in thyroid tumorigenesis, where PBF is generally overexpressed and p53 mutations relatively rare. It will also be important to determine whether high thyroidal PBF is a risk factor for irradiation-associated tumors, especially in individuals with multinodular goiters in which elevated PBF expression has been described (11). Together these findings further emphasize that PBF has an increasingly important role in the etiology of thyroid disease.
Supplementary Material
Acknowledgments
We thank Dean Gentle, Andrea Bacon, Ann Logan and Wendy Leadbeater for technical help. This work was supported by grants from the Get-Ahead Charity, the Medical Research Council, the Wellcome Trust and Cancer Research UK.
Footnotes
Disclosure Summary: The authors have nothing to disclose.
References
- 1.Chen AY, Jemal A, Ward EM. Increasing Incidence of Differentiated Thyroid Cancer in the United States, 1988–2005. Cancer. 2009;115:3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 2.Bhaijee F, Nikiforov YE. Molecular Analysis of Thyroid Tumors. Endocr Pathol. 2011;22:126–133. doi: 10.1007/s12022-011-9170-y. [DOI] [PubMed] [Google Scholar]
- 3.Mazzaferri EL, Jhiang SM. Long-Term Impact of Initial Surgical and Medical Therapy on Papillary and Follicular Thyroid-Cancer. Am J Med. 1994;97:418–428. doi: 10.1016/0002-9343(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 4.Nixon IJ, Ganly I, Patel SG, Palmer FL, Di Lorenzo MM, Grewal RK, Larson SM, Tuttle RM, Shaha A, Shah JP. The Results of Selective Use of Radioactive Iodine on Survival and on Recurrence in the Management of Papillary Thyroid Cancer, Based on Memorial Sloan-Kettering Cancer Center Risk Group Stratification. Thyroid. 2013;23:683–694. doi: 10.1089/thy.2012.0307. [DOI] [PubMed] [Google Scholar]
- 5.Xing MZ, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, Yip L, Mian C, Vianello F, Tuttle RM, Robenshtok E, Fagin JA, Puxeddu E, Fugazzola L, Czarniecka A, Jarzab B, O’Neill CJ, Sywak MS, Lam AK, Riesco-Eizaguirre G, Santisteban P, Nakayama H, Tufano RP, Pai SI, Zeiger MA, Westra WH, Clark DP, Clifton-Bligh R, Sidransky D, Ladenson PW, Sykorova V. Association Between BRAF V600E Mutation and Mortality in Patients With Papillary Thyroid Cancer. Jama-J Am Med Assoc. 2013;309:1493–1501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien W, Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J Biol Chem. 2000;275:19422–19427. doi: 10.1074/jbc.M910105199. [DOI] [PubMed] [Google Scholar]
- 7.Stratford AL, Boelaert K, Tannahill LA, Kim DS, Warfield A, Eggo MC, Gittoes NJL, Young LS, Franklyn JA, McCabe CJ. Pituitary tumor transforming gene binding factor: A novel transforming gene in thyroid tumorigenesis. J Clin Endocr Metab. 2005;90:4341–4349. doi: 10.1210/jc.2005-0523. [DOI] [PubMed] [Google Scholar]
- 8.Boelaert K, Smith VE, Stratford AL, Kogai T, Tannahill LA, Watkinson JC, Eggo MC, Franklyn JA, McCabe CJ. PTTG and PBF repress the human sodium iodide symporter. Oncogene. 2007;26:4344–4356. doi: 10.1038/sj.onc.1210221. [DOI] [PubMed] [Google Scholar]
- 9.Smith VE, Read ML, Turnell AS, Watkins RJ, Watkinson JC, Lewy GD, Fong JCW, James SR, Eggo MC, Boelaert K, Franklyn JA, McCabe CJ. A novel mechanism of sodium iodide symporter repression in differentiated thyroid cancer. Journal of Cell Science. 2009;122:3393–3402. doi: 10.1242/jcs.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith VE, Read ML, Turnell AS, Sharma N, Lewy GD, Fong JCW, Seed RI, Kwan P, Ryan G, Mehanna H, Chan SY, Darras VM, Boelaert K, Franklyn JA, McCabe CJ. PTTG-Binding Factor (PBF) Is a Novel Regulator of the Thyroid Hormone Transporter MCT8. Endocrinology. 2012;153:3526–3536. doi: 10.1210/en.2011-2030. [DOI] [PubMed] [Google Scholar]
- 11.Read ML, Lewy GD, Fong JCW, Sharma N, Seed RI, Smith VE, Gentilin E, Warfield A, Eggo MC, Knauf JA, Leadbeater WE, Watkinson JC, Franklyn JA, Boelaert K, McCabe CJ. Proto-oncogene PBF/PTTG1IP Regulates Thyroid Cell Growth and Represses Radioiodide Treatment. Cancer Research. 2011;71:6153–6164. doi: 10.1158/0008-5472.CAN-11-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsueh C, Lin J, Chang Y, Hsueh S, Chao T, Yu J, Jung S, Tseng N, Sun J, SY K, SH U. Prognostic significance of pituitary tumor-transforming gene-binding factor (PBF) expression in papillary thyroid carcinoma. Clin Endocrinol (Oxf) 2012;78:303–309. doi: 10.1111/cen.12007. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Human Mutation. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 14.Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Borresen-Dale AL. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 15.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smallridge RC, Marlow LA, Copland JA. Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocr-Relat Cancer. 2009;16:17–44. doi: 10.1677/ERC-08-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malaguarnera R, Vella V, Vigneri R, Frasca F. p53 family proteins in thyroid cancer. Endocr Relat Cancer. 2007;14:43–60. doi: 10.1677/erc.1.01223. [DOI] [PubMed] [Google Scholar]
- 18.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, Helin K, Pelicci PG, Jochemsen AG, Marine JC. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayburn E, Zhang RW, He J, Wang H. MDM2 and human malignancies: Expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Tar. 2005;5:27–41. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- 20.Jennings T, Bratslavsky G, Gerasimov G, Troshina K, Bronstein M, Dedov I, Alexandrova G, Figge J. Nuclear accumulation of MDM2 protein in well-differentiated papillary thyroid carcinomas. Exp Mol Pathol. 1995;62:199–206. doi: 10.1006/exmp.1995.1022. [DOI] [PubMed] [Google Scholar]
- 21.Zou MJ, Shi YF, Alsedairy S, Hussain SS, Farid NR. The Expression of the Mdm2 Gene, a P53 Binding-Protein, in Thyroid Carcinogenesis. Cancer. 1995;76:314–318. doi: 10.1002/1097-0142(19950715)76:2<314::aid-cncr2820760223>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 22.Frasca F, Rustighi A, Malaguarnera R, Altamura S, Vigneri P, Del Sal G, Giancotti V, Pezzino V, Vigneri R, Manfioletti G. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Research. 2006;66:2980–2989. doi: 10.1158/0008-5472.CAN-05-2637. [DOI] [PubMed] [Google Scholar]
- 23.Esposito F, Tornincasa M, Federico A, Chiappetta G, Pierantoni GM, Fusco A. High-mobility group A1 protein inhibits p53-mediated intrinsic apoptosis by interacting with Bcl-2 at mitochondria. Cell Death Dis. 2012:e383. doi: 10.1038/cddis.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Imaizumi M, Usa T, Tominaga T, Neriishi K, Akahoshi M, Nakashima E, Ashizawa K, Hida A, Soda M, Fujiwara S, Yamada M, Ejima E, Yokoyama N, Okubo M, Sugino K, Suzuki G, Maeda R, Nagataki S, Eguchi K. Radiation dose-response relationships for thyroid nodules and autoimmune thyroid diseases in Hiroshima and Nagasaki Atomic Bomb survivors 55–58 years after radiation exposure. Jama-J Am Med Assoc. 2006;295:1011–1022. doi: 10.1001/jama.295.9.1011. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X. Tied Up in Loops: Positive and Negative Autoregulation of p53. Csh Perspect Biol. 2010;2:a000984. doi: 10.1101/cshperspect.a000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet. 2002;32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- 28.Yu R, Heaney AP, Lu W, Chen J, Melmed S. Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. J Biol Chem. 2000;275:36502–36505. doi: 10.1074/jbc.C000546200. [DOI] [PubMed] [Google Scholar]
- 29.Messina RL, Sanfilippo M, Vella V, Pandini G, Vigneri P, Nicolosi ML, Giani F, Vigneri R, Frasca F. Reactivation of p53 mutants by p53 reactivation and induction of massive apoptosis in thyroid cancer cells. Int J Cancer. 2012;130:2259–2270. doi: 10.1002/ijc.26228. [DOI] [PubMed] [Google Scholar]
- 30.Wyllie FS, Haughton MF, Rowson JM, Wynford-Thomas D. Human thyroid cancer cells as a source of iso-genic, iso-phenotypic cell lines with or without functional p53. Brit J Cancer. 1999;79:1111–1120. doi: 10.1038/sj.bjc.6690177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blagosklonny MV, Giannakakou P, Wojtowicz M, Romanova LY, Ain KB, Bates SE, Fojo T. Effects of p53-expressing adenovirus on the chemosensitivity and differentiation of anaplastic thyroid cancer cells. J Clin Endocr Metab. 1998;83:2516–2522. doi: 10.1210/jcem.83.7.4984. [DOI] [PubMed] [Google Scholar]
- 32.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland JA, Smallridge RC, Haugen BR. Deoxyribonucleic Acid Profiling Analysis of 40 Human Thyroid Cancer Cell Lines Reveals Cross-Contamination Resulting in Cell Line Redundancy and Misidentification. J Clin Endocr Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins RJ, Read ML, Smith VE, Sharma N, Reynolds GM, Buckley L, Doig C, Campbell MJ, Lewy G, Eggo MC, Loubiere LS, Franklyn JA, Boelaert K, McCabe CJ. Pituitary tumor transforming gene binding factor: a new gene in breast cancer. Cancer Res. 2010;70:3739–3749. doi: 10.1158/0008-5472.CAN-09-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2009;11:36–42. [Google Scholar]
- 35.Rasti m, Grand RJ, Yousef YF, Shuen M, Mymryk JS, Gallimore PH, Turnell AS. Roles for the APIS and the 20S proteasome in adenovirus E1A-dependent transcription. EMBO. 2006;25:2710–2722. doi: 10.1038/sj.emboj.7601169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sjogren S, Inganas M, Norberg T, Lindgren A, Nordgren H, Holmberg L, Bergh J. The p53 gene in breast cancer: Prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer I. 1996;88:173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- 37.Kim DS, Franklyn JA, Smith VE, Stratford AL, Pemberton HN, Warfield A, Watkinson JC, Ishmail T, Wakelam MJ, McCabe CJ. Securin induces genetic instability in colorectal cancer by inhibiting double-stranded DNA repair activity. Carcinogenesis. 2007;28:749–759. doi: 10.1093/carcin/bgl202. [DOI] [PubMed] [Google Scholar]
- 38.Basik M, Stoler DL, Kontzoglou KC, RodriguezBigas MA, Petrelli NJ, Anderson GR. Genomic instability in sporadic colorectal cancer quantitated by inter-simple sequence repeat PCR analysis. Gene Chromosome Canc. 1997;18:19–29. [PubMed] [Google Scholar]
- 39.Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- 40.Smith VE, Sharma N, Watkins RJ, Read ML, Ryan G, Kwan P, Watkinson JC, Boelaert K, Franklyn JA, McCabe CJ. Manipulation of PBF/PTTG1IP phosphorylation status: a potential new therapeutic strategy for improving radioiodine uptake in thyroid and other tumors. J Clin Endocrinol Metab. 2013;98:2876–2886. doi: 10.1210/jc.2012-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Wang DL, Liu Y, Zhao L, Sun FL. RAD6 Regulates the Dosage of p53 by a Combination of Transcriptional and Posttranscriptional Mechanisms. Molecular and Cellular Biology. 2012;32:576–587. doi: 10.1128/MCB.05966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shekhar MPV, Lyakhovich A, Visscher DW, Heng H, Kondrat N. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Research. 2002;62:2115–2124. [PubMed] [Google Scholar]
- 43.Yaspo ML, Aaltonen J, Horelli-Kuitunen N, Peltonen L, Lehrach H. Cloning of a novel human putative type Ia integral membrane protein mapping to 21q22.3. Genomics. 1998;49:133–136. doi: 10.1006/geno.1998.5217. [DOI] [PubMed] [Google Scholar]
- 44.McCabe CJ, Khaira JS, Boelaert K, Heaney AP, Tannahill LA, Hussain S, Mitchell R, Olliff J, Sheppard MC, Franklyn JA, Gittoes NJL. Expression of pituitary tumour transforming gene (PTTG) and fibroblast growth factor-2 (FGF-2) in human pituitary adenomas: relationships to clinical tumour behaviour. Clin Endocrinol. 2003;58:141–150. doi: 10.1046/j.1365-2265.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 45.Yuan J, Luo KT, Zhang LZ, Cheville JC, Lou ZK. USP10 Regulates p53 Localization and Stability by Deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Wijk SJL, Timmers HTM. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. Faseb Journal. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 47.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez MS, Desterro JMP, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Molecular and Cellular Biology. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, Byeon IJL, Gabizon R, Mattia M, Zupnick A, Brown LM, Friedler A, Prives C. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol. 2010;17:982–995. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. Embo Journal. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Molecular and Cellular Biology. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Y, Luo JY, Zhang WZ, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Molecular Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 53.Sui GC, El Bachir A, Shi YJ, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Kim DS, Fong J, Read ML, McCabe CJ. The emerging role of pituitary tumour transforming gene (PTTG) in endocrine tumouri-genesis. Mol Cell Endocrinol. 2007;278:1–6. doi: 10.1016/j.mce.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, Fukuzawa M, Nakagawara A. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. Journal of Biological Chemistry. 2004;279:25549–25561. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- 56.Grombacher T, Eichhorn U, Kaina B. P53 is involved in regulation of the DNA repair gene O-6-methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene. 1998;17:845–851. doi: 10.1038/sj.onc.1202000. [DOI] [PubMed] [Google Scholar]
- 57.Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D’Erchia AM, Navarro B, Tullo A, Saccone C, Gisel A. P53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. Bmc Bioinformatics. 2007;8:S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability - an evolving hallmark of cancer. Nat Rev Mol Cell Bio. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 59.Bastos HN, Antao MR, Silva SN, Azevedo AP, Manita I, Teixeira V, Pina JE, Gil OM, Ferreira TC, Limbert E, Rueff J, Gaspar JF. Association of Polymorphisms in Genes of the Homologous Recombination DNA Repair Pathway and Thyroid Cancer Risk. Thyroid. 2009;19:1067–1076. doi: 10.1089/thy.2009.0099. [DOI] [PubMed] [Google Scholar]
- 60.de Vogel S, Weijenberg MP, Herman JG, Wouters KAD, de Goeij AFPM, van den Brandt PA, de Bruine AP, van Engeland M. MGMT and MLH1 promoter methylation versus APC, KRAS and BRAF gene mutations in colorectal cancer: indications for distinct pathways and sequence of events. Ann Oncol. 2009;20:1216–1222. doi: 10.1093/annonc/mdn782. [DOI] [PubMed] [Google Scholar]
- 61.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1199. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.