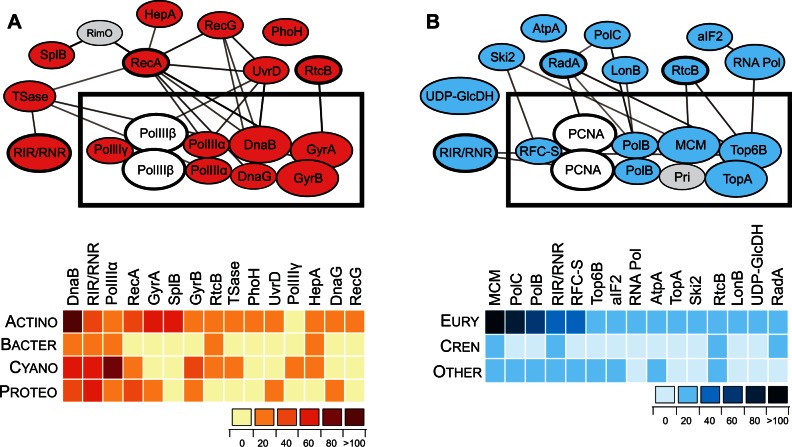
Fig. 4.
Intein-containing proteins are often members of the same complexes and networks. (A) Top 15 bacterial intein-containing proteins, their interactions, and intein distribution. Proteins of the DNA replication fork are boxed at the center of the network. Network was reconstructed using STRING database of known and predicted protein interactions (http://string-db.org/; Szklarczyk et al. 2015). The list of the proteins, their full names, and description is available in table 1. Critical proteins with no inteins are shown in gray as follows: PolIIIβ, DNA polymerase III beta subunit; RimO, 2-methylthioadenine synthetase. The heatmap reflects distribution of the inteins among listed proteins (top) of four bacterial clades (side): Actinobacteria (Actino), Bacteroidetes (Bacter), Cyano (Cyanobacteria), and Proteobacteria (Proteo). For full network and list of the proteins, see supplementary figure S3 and table S5, Supplementary Material online. (B) Top 15 archaeal intein-containing proteins, their interactions, and intein distribution. Proteins of the DNA replication fork are boxed at the center of the network. Network was reconstructed as in panel (A) and the proteins listed in table 2. Critical proteins with no inteins are shown in gray as follows: PCNA, proliferating cell nuclear antigen or DNA clamp; Pri, primase. The heatmap is displayed as in (A) with three groups indicated on the side: Euryarchaeota (Eury), Crenarchaeota (Cren), and other archaea (Other). For full network and list of the proteins, see supplementary figure S4 and table S6, Supplementary Material online.