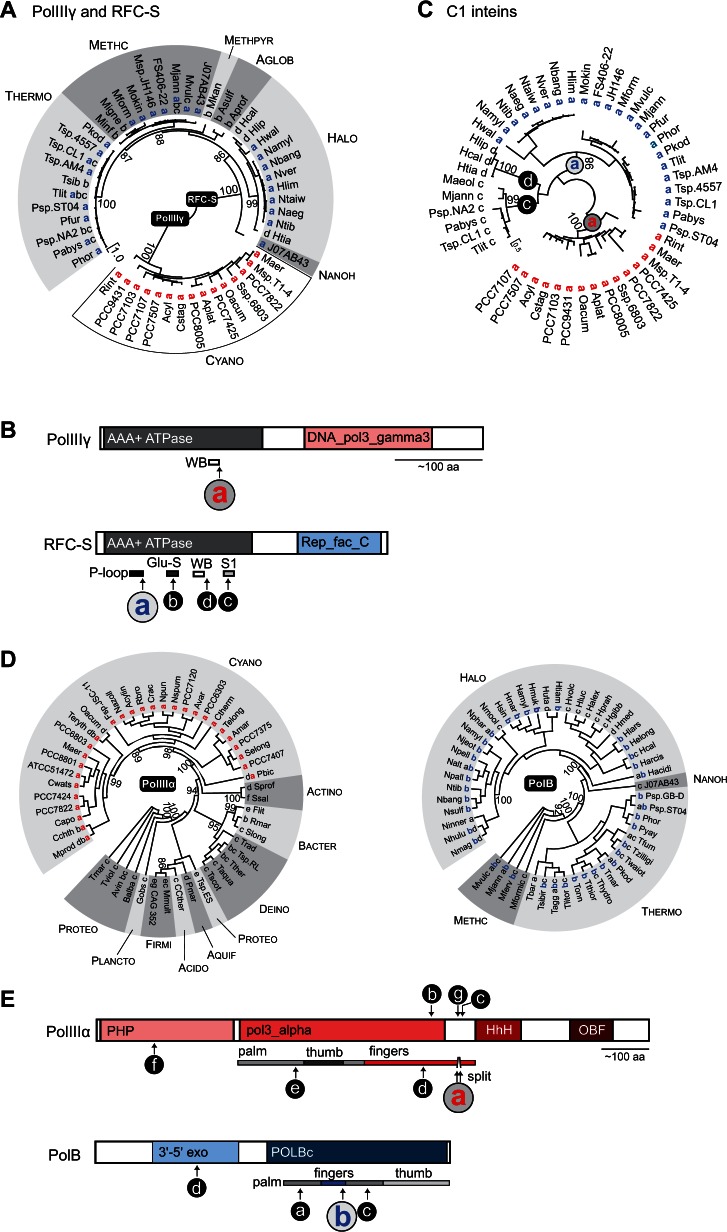
Fig. 6.
Inteins in bacterial and archaeal clamp loaders and DNA polymerases cluster in functional domains. (A) Distribution of inteins in PolIIIγ and RFC-S. Phylogenetic tree for ATPase domain of the clamp loader proteins from both bacteria and archaea was reconstructed based on the amino acid sequences using the ML algorithm with WAG model. Statistical support for the tree was evaluated with SH-aLRT; however, only values for critical nodes, which were higher than 85%, are shown. The intein insertion point(s) a–d and abbreviated species names are shown next to branches. Letters for insertion points in PolIIIγ and RFC-S do not correspond to each other (see B and C). The tree with full-length species names is available in supplementary figure S9, Supplementary Material online; the trees reconstructed based on the extended data sets including intein-containing and intein-less proteins are available in supplementary figures S10 and S11, Supplementary Material online. PolIIIγ inteins were found only in Cyanobacteria (Cyano). Archaeal clades as follows: Thermo, Thermococci; Methc, Methanococci; Methpyr, Methanopyri; Nanoh, Nanohaloarchaeota; Halo, Halobacteria; Aglob, Archaeolglobi. (B) Intein insertion points. Intein locations are shown along PolIIIγ and RFC-S relative to structural and functional domains. The ATPase domain (AAA+ ATPase, black) has a single intein insertion in PolIIIγ (site a shown in red) and multiple intein insertion points in RFC-S (sites a–d). The insertion point a in PolIIIγ is located in highly conserved Walker B motif (WB). The most common insertion point a in RFC-S (blue) is located in P-loop. Other motifs shown for RFC-S are: Glu-S, glutamine switch; S1, sensor one. PolIIIγ and RFC-S proteins have additional domains specific for respective proteins: DNA_pol3_gamma3 domain (pink) is found only in PolIIIγ, whereas Rep_fac_C domain (light blue) is present only in RFC-S proteins. (C) Phylogenetic analysis of the C1 inteins from PolIIIγ and RFC-S. Phylogenetic tree was reconstructed based on the intein splicing domain amino acid sequences using the ML algorithm with WAG model. Statistical support for the tree was evaluated with SH-aLRT; however, only values for critical nodes, which were higher than 85%, are shown. Only inteins with cysteine as the first amino acid residue (C1 inteins) were used, which included all inteins identified in PolIIIγ (insertion point a, red), and inteins from insertion points a (blue), c, and d from RFC-S. The intein insertion point(s) a, c, d and abbreviated species names are shown next to branches. The intein insertion point(s) are also indicated in the nodes. The tree with full-length species names is available in supplementary figure S12, Supplementary Material online. (D) Distribution of inteins in PolIIIα and PolB. Phylogenetic trees for bacterial replicative DNA polymerase PolIIIα and archaeal PolB were reconstructed based on the extein amino acid sequences using the ML algorithm with WAG model. Statistical support was evaluated with SH-aLRT; however, only values for critical nodes, which were higher than 85%, are shown. The intein insertion point(s) a–f and abbreviated species names are shown next to branches. Letters for insertion points in PolIIIα and PolB do not correspond to each other (see B). The full-length trees with full-length species names are available in supplementary figures S13 and S14, Supplementary Material online. Although PolIIIα and PolB are functionally equivalent counterparts in bacteria and archaea, these proteins are not related. Bacterial clades as follows: Cyano, Cyanobacteria; Actino, Actinobacteria; Bacter, Bacteroidetes; Deino, Deionococcus–Thermus; Acido, Acidobacteria; Plancto, Planctomycetes; Proteo, Proteobacteria; Aquif, Aquificae; and Firmi, Firmicutes. Archaeal clades as follows: Halo, Halobacteria; Nanoh, Nanohaloarchaeota; Methc, Methanococci; Thermo, Thermococci. (E) Intein insertion points. Intein locations are shown along PolIIIα and PolB relative to structural and functional domains. The critical catalytic domains have multiple intein insertion points in both PolIIIα (pol3_alpha) and PolB (POLBc). Additional insertion points were found in bacterial PHP (polymerase and histidinol hhosphatase domain) for PolIIIα and in archaeal 3′–5′ exo (3′–5′ exonuclease domain of archaeal family-B DNA polymerases) for PolB. Polymerase structural domains are shown on the bottom. PolIIIα inteins from insertion point a are split. Additional abbreviations: HhH, helix-hairpin-helix DNA-binding domain; OBF, (oligonucleotide/oligosaccharide binding)-fold.