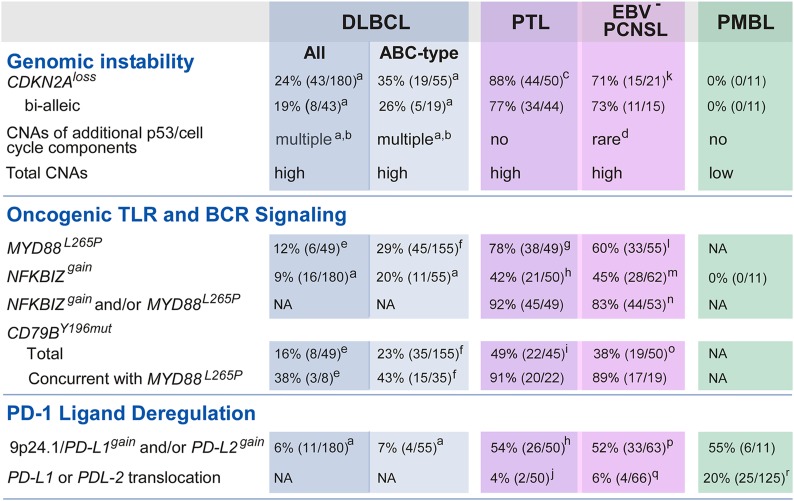
Figure 7.
Unique combinations of structural alterations in discrete LBCL subtypes. Frequency of specific genetic alterations modulating “Genomic Instability,” “Oncogenic TLR and BCR Signaling,” and “PD-1 Ligand Deregulation” in DLBCL all, DLBCL ABC-type, PTL, EBV– PCNSL, and PMBL are noted. a, Raw data previously published in reference 14. b, CNAs include the following alterations: MDM2gain, MDM4gain, CDK2gain, CDK4gain, CDK6gain, RB1loss, RBL2loss, TP53loss, KDM6Bloss, RPL26loss, BCL2L12gain, RFWD2gain, CCND3gain. c, CDKN2A CN data were available for 50 PTL (7 discovery + 43 extension). d, Only RBL2loss. e, As reported in reference 54. f, As reported in reference 8. g, MYD88L265P mutation status was available from 49 PTL (7 discovery + 42 extension). h, CN data for NFKBIZ and 9p24.1/PD-L1/PD-L2 loci were available from 50 PTL (7 discovery + 43 extension). i, CD79BY196mut mutation status in 45 PTL (7 discovery + 38 extension). j, 9p24.1/PD-L1/PD-L2 translocation data were from 50 PTL (7 discovery + 43 extension). k, CDKN2A CN data were available for 21 EBV– PCNSL (discovery only). l, MYD88L265P mutation status was available from 55 EBV– PCNSL (14 discovery + 41 extension). m, CN data for NFKBIZ locus were available from 62 EBV– PCNSL (21 discovery + 41 extension). n, NFKBIZ CN data and MYD88L265P mutation status were available for 53 EBV– PCNSL (12 discovery + 41 extension). o, CD79BY196mut mutation status was available from 50 EBV– PCNSL (12 discovery + 38 extension). p, 9p24.1/PD-L1/PD-L2 CN data were from 63 EBV– PCNSL (21 discovery + 42 extension). q, 9p24.1/PD-L1/PD-L2 translocation data were from 66 EBV– PCNSL (24 discovery + 42 extension). r, as reported in reference 19.