Summary
Human CD4+CD25hiFoxp3+CD127− Treg and CD4+CD25−Foxp3− Tconv cell functions are governed by their metabolic requirements. Here we report a comprehensive comparative analysis between ex vivo human Treg and Tconv cells that comprises analyses of the proteomic networks in subcellular compartments. We identified a dominant proteomic signature at the metabolic level that primarily impacted the highly-tuned balance between glucose and fatty-acid oxidation in the two cell types. Ex vivo Treg cells were highly glycolytic while Tconv cells used predominantly fatty-acid oxidation (FAO). When cultured in vitro, Treg cells engaged both glycolysis and FAO to proliferate, while Tconv cell proliferation mainly relied on glucose metabolism. Our unbiased proteomic analysis provides a molecular picture of the impact of metabolism on ex vivo human Treg versus Tconv cell functions that might be relevant for therapeutic manipulations of these cells.
Keywords: Metabolism, Regulatory T cells, Conventional T cells, Proteomic Analysis, Immune Tolerance
Graphical Abstract
Highlights
-
•
Ex vivo human Treg cells are highly glycolytic and proliferating
-
•
Ex vivo human Tconv cells use fatty-acid oxidation (FAO) and are non-proliferating
-
•
In vitro proliferation of human Treg cells requires both glycolysis and FAO
-
•
In vitro proliferation of human Tconv cells relies mainly on glycolysis
The proteomic signature of human Treg and Tconv cells and their functional specialization is poorly understood. Matarese and colleagues identified metabolic pathway signatures that suggest a highly-tuned balance between glucose and fatty-acid oxidation impacting functions of in vitro versus ex vivo human Treg and Tconv cells.
Introduction
CD4+ T cells have been classified into different functionally distinct subsets, on the basis of their cytokine production patterns that generally associate with the expression of multiple lineage-specific transcription factors (Bluestone et al., 2009). Among those factors, the forkhead-box-P3 (FoxP3) transcription factor is expressed by CD4+CD25+ regulatory T (Treg) cells, a specialized subset of CD4+ T cells that suppresses proliferation and effector cell functions in a wide range of immune target cells (Sakaguchi et al., 2008, Min et al., 2003, Zheng et al., 2004, Kohrt et al., 2010, Khazaie and von Boehmer, 2006). Human Treg cells display a series of “apparent paradoxes” in their immunobiology: they manifest in vitro hyporesponsiveness (anergy) to T cell receptor (TCR) stimulation (Thornton and Shevach, 1998, Li et al., 2005) although they have high surface expression of activation markers and are highly proliferative in vivo (Vukmanovic-Stejic et al., 2006, Vukmanovic-Stejic et al., 2008). In contrast, CD4+CD25−FoxP3− conventional T (Tconv) cells are not hyporesponsive to TCR stimulation in vitro, but rapidly respond to antigenic stimulation by increasing production of interleukin-2 (IL-2) and/or cytokines that sustain their own proliferation and clonal differentiation toward effector phenotypes.
Treg and Tconv cells have a high degree of plasticity that associates with a different regulation of their own transcriptional programs. Over the past few years, advances have been made in the understanding of the transcriptional regulation underlying the gene-expression profiles of these cells (Schmidl et al., 2014, Luo and Li, 2013, Painter et al., 2011). In particular, the integration of multiple extracellular signals that directly affect transcriptional programs and signaling pathways in both cellular subsets have been linked to the induction of proliferation, production of cytokines, and modulation of energy metabolism. In this report, we mapped the proteome of either freshly-isolated, in vitro-cultured, or TCR-activated human Treg and Tconv cells to dissect their biochemical and metabolic profiles and evaluate their dynamic changes upon different in vitro culture conditions. Because the functions of Treg and Tconv cells are regulated by specific metabolic pathways, the full understanding of how they change according to specific microenvironmental conditions and energy demands could have major implications in integrative pathophysiology and human autoimmunity.
Results
Freshly-Isolated Human Treg Cells Are Glycolytic, whereas Tconv Cells Use Fatty-Acid Oxidation
The proteomic landscape of human Treg and Tconv cells was assessed by analyzing protein expression according to their subcellular compartmentalization (either cytosolic- or membrane-associated). Highly stringent criteria in raw data handling ensured the confident identification of 6,610 distinct peptides, corresponding overall to 1,860 unique proteins. According to the theoretical molecular weight (MW) and isoelectric point (pI), the proteins identified were plotted in a 2D map using the Multidimensional Algorithm Protein Map (MAProMa) software (Brambilla et al., 2012) (data not shown). The identified proteins, including those differentially represented, were plotted into the Global Mammalian Protein Interactomic (GMPI) network, using the Cytoscape platform and its plugins (see Supplemental Experimental Procedures for details).
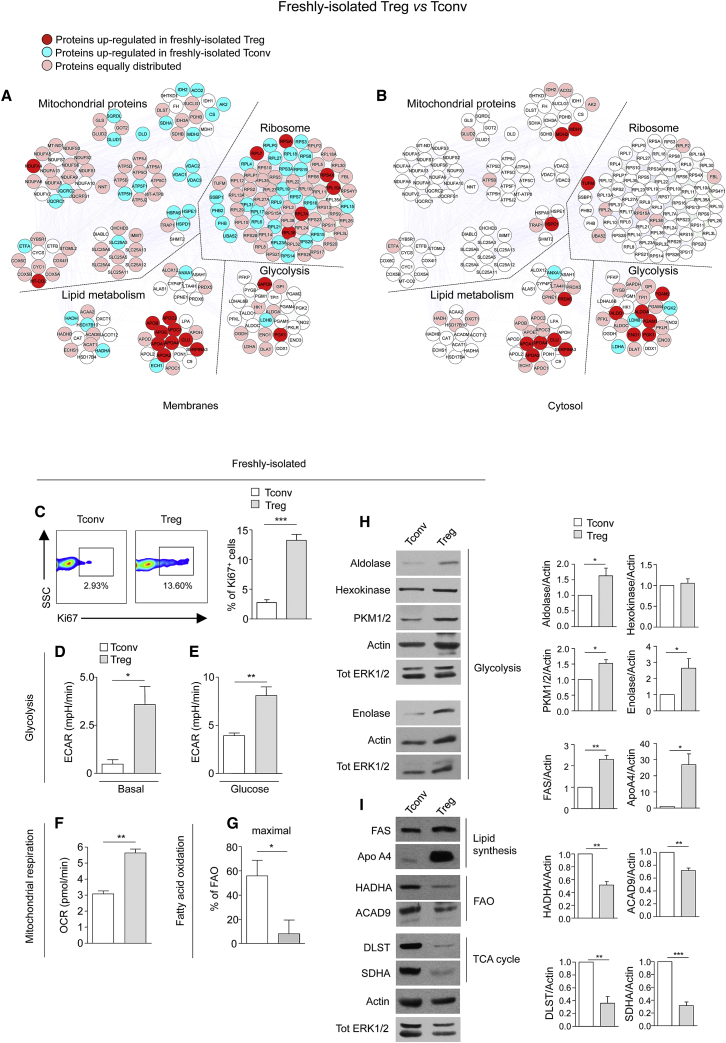
To delineate the basal proteomic signature and networks of human Treg and Tconv cells, we compared protein-expression profiles of the two freshly-isolated cell subsets, using Differential Average (Dave) and Differential Coefficient Index (DCI) algorithms from MAProMa (Mauri et al., 2005). The differentially expressed proteins are listed in Table S1A, Figure S1A, Table S1B, and Figure S1B (membranes and cytosol, respectively). Because the most representative functional classes that we had found differentially expressed between freshly-isolated Treg and Tconv cells were those associated with metabolism, we analyzed this aspect at the protein, biochemical, and functional levels. Proteomic analysis of freshly-isolated human Treg cells indicated an upregulation of glycolysis-related proteins (Figures 1A and 1B and Table S1C), such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoglycerate kinase 1 (PGK1) (in membranes) and transaldolase 1 (TALDO1), aldolase A (ALDOA), phosphoglycerate mutase 1 and 2 (PGAM1 and 2), enolase 1 (ENO1), and PGK1 (in the cytosol), in agreement with the high proliferative profile of these cells in vivo (Vukmanovic-Stejic et al., 2006, Vukmanovic-Stejic et al., 2008). In contrast, Tconv cells expressed higher amounts of mitochondrial proteins than Treg cells including enzymes of the Krebs tricarboxylic acid cycle (TCA) (isocitrate dehydrogenase [IDH2], aconitase 2 [ACO2], citrate synthase [CS], malate dehydrogenase [MDH2], succinate dehydrogenase [SDHA]), proteins involved in the mitochondrial respiratory electron transport chain (electron transfer flavoprotein [ETFA], ubiquinol-cytochrome c reductase I and II [UQCRC1 and 2]), in the anion channel formation (voltage-dependent anion channel 1, 2 and 3 [VDAC1, 2, and 3]), in the control of mitochondrial permeability (solute carrier family 25 [SLC25A3]), and also mitochondrial proteins devoted to production of ATP (H+-ATP synthase subunit b [ATP5F1], ATP synthase H+ transporting [ATP5H]). In addition, Treg cells had consistently higher amount of lipid metabolism-associated proteins than Tconv cells, including apolipoprotein B-100 (Apo B), apolipoprotein E (ApoE), apolipoprotein C-III precursor (APOC3), apolipoprotein A-I preproprotein (APOA1), apolipoprotein A-IV precursor (APOA4), apolipoprotein C-II precursor (APOC2), apolipoprotein A-II preproprotein (APOA2), clusterin isoform 2 preproprotein (CLU), and serpin peptidase inhibitor member 3 (Serpin A3).
Figure 1.
Freshly-Isolated Human Treg Cells Display a Glycolytic Metabolism whereas Tconv Cells Preferentially Use FAO
(A and B) Map of the interactome networks controlling the main biological processes/functions associated to metabolism (mitochondrial proteins, ribosome, lipid metabolism and glycolysis), obtained by comparing the protein profile of freshly-isolated Tconv and Treg cells in the membranes (A) and in the cytoplasm (B). Red plots correspond to specific proteins upregulated in Treg cells; blue plots correspond to proteins upregulated in Tconv cells; pink plots represent the equally distributed proteins in the two cell compartments; white plots are the not-identified proteins.
(C) Left, representative flow cytometry plots (percentage of Ki67+ cells indicated above the boxes) from freshly-isolated Tconv and Treg cells (SSC, side scatter) (one representative plot out of ten). Right shows percentage of Ki67+ cells from freshly-isolated Tconv and Treg cells. The data are shown as mean ± SEM of ten independent experiments.
(D and E) ECAR in basal condition (D) and upon glucose injection (E) on freshly-isolated Tconv and Treg cells (one representative out of three independent experiments). The data are shown as mean ± SEM of three measurements, each of them in duplicates.
(F) OCR in freshly-isolated Tconv and Treg cells (one representative out of three independent experiments). The data are shown as mean ± SEM of three measurements, each of them in duplicates.
(G) Percentage of FAO in freshly-isolated Tconv and Treg cells (one representative out of three independent experiments). The data are shown as mean ± SEM of three measurements, each of them in triplicates. (C–G) Statistical analysis by two tailed Mann-Whitney test. (∗p < 0.05; ∗∗p < 0.005, ∗∗∗p < 0.0005).
(H and I) Immunoblot for aldolase, enolase, hexokinase, PKM1/2, FAS, ApoA4, HADHA, ACAD9, DLST, and SDHA on freshly-isolated Tconv and Treg cells. Actin and total ERK 1/2 served as a loading control. One representative out of at least two independent experiments is shown. The graphs show the relative densitometric quantitation of aldolase, hexokinase, PKM1/2, enolase, FAS, ApoA4, HADHA, ACAD9, DLST, and SDHA normalized on actin in freshly-isolated Tconv and Treg cells and shown as fold over Tconv cells (n = 6; data are shown as mean ± SEM of two independent experiments, in triplicates). (H–I) Statistical analysis by paired two-tailed Student’s t test (∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001).
To assess whether the proteomic signature associated with specific differences of cellular metabolism, we performed functional analyses (Seahorse assays), evaluating the bioenergetic profiles of freshly-isolated Treg and Tconv cells. We confirmed that Treg cells ex vivo were more proliferative than Tconv cells, as testified by an increased expression of Ki67+cells in the former compartment (Figure 1C). Next, we measured the extracellular acidification rate (ECAR), an indicator of aerobic glycolysis, in freshly-isolated Treg and Tconv cells, and found that Treg cells had higher basal and glucose-stimulated ECAR (Figures 1D and 1E) than Tconv cells. In parallel, we found that O2 consumption rate (OCR), an indicator of oxidative phosphorylation (OXPHOS), was higher in Treg as compared to Tconv cells, suggesting an increased mitochondrial functionality that might possibly associate with an enhanced capacity of Treg cells to adapt to an increased metabolic demand (Figure 1F). Finally, fatty-acid β-oxidation (FAO) was evaluated by measuring OCR resulting from exogenous addition of fatty acids (palmitate) to cells, in the presence or in the absence of etomoxir (a specific inhibitor of FAO), to discriminate the fraction of OCR associated with fatty acid utilization versus that derived from other substrates. Tconv cells displayed higher FAO than Treg cells, suggesting that non-proliferating cells mainly use FAO as an energy source (Figures 1G and S1C).
These data were confirmed at the biochemical level (Figures 1H and 1I and S1D–S1G). The expression of glycolysis-related enzymes (enolase, aldolase, pyruvate kinase 1/2 [PKM1/2]), as well as lipid metabolism-related molecules (ApoA4 and fatty acid synthase, FAS) was upregulated in Treg as compared to Tconv cells. On the contrary, Tconv cells had an increased expression of enzymes involved in FAO (hydroxyacyl-Coenzyme A dehydrogenase [HADHA], and acyl-CoA dehydrogenase [ACAD9]) and the TCA cycle (dihydrolipoamide succinyltransferase [DLST] and succinate dehydrogenase [SDHA]).
Distinct Proteomic and Metabolic Profiles of In Vitro Anergic versus Proliferating Human Treg Cells
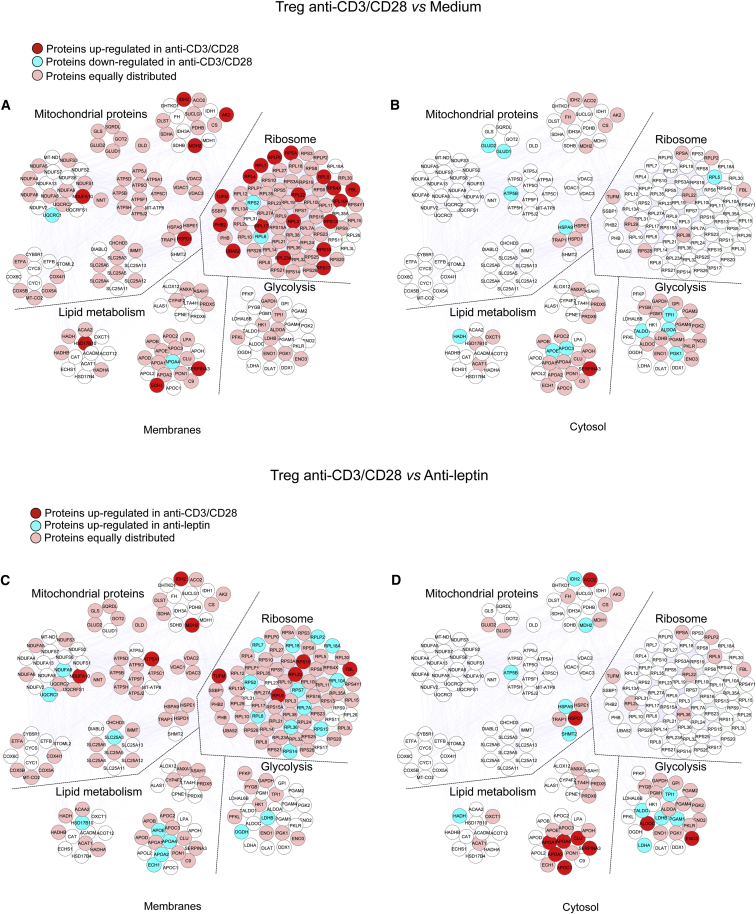
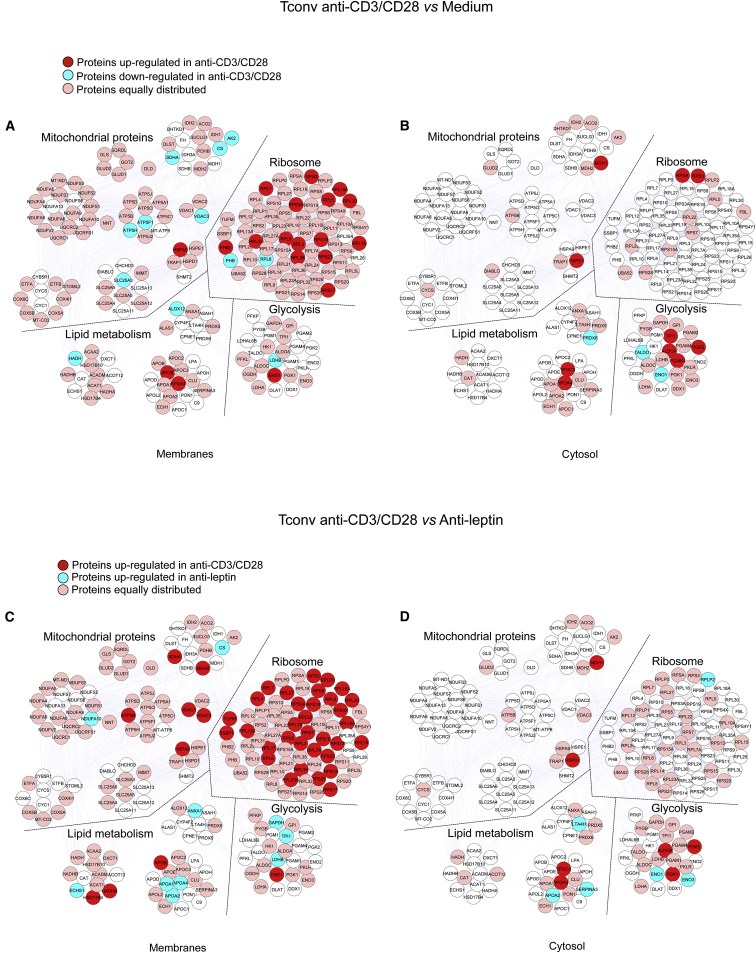
Treg cells proliferate in vivo (Vukmanovic-Stejic et al., 2006, Vukmanovic-Stejic et al., 2008) despite their anergic state in vitro (Li et al., 2005). The reason for this discrepancy is little understood, although metabolism seems to play a role in this process, as indicated by the evidence that overactivation of the mammalian target of rapamycin (mTOR) pathway associates with human Treg cell anergy (Procaccini et al., 2010, Procaccini and Matarese, 2012). To determine whether in vitro culture conditions could affect metabolic programs, we analyzed the proteomic profile of Treg cells after 12 hr of culture in the presence or in the absence of TCR-mediated stimulation with anti-CD3 and anti-CD28 (Tables S2A–S2C and Figures S2A and S2B). Proteomic analysis of in vitro (anergic) Treg cells revealed that in the membrane compartment there was an unbalanced regulation of mitochondrial proteins upon TCR stimulation (Figure 2A and Table S2C). Indeed, IDH2, adenylate kinase 2 (AK2) (involved in energy metabolism), MDH2 (citric acid cycle), NADH dehydrogenase 1 α subcomplex 10 (NDUFA10) (electron transfer), and mitochondrial heat shock protein 60 (HSPD1) were upregulated by TCR stimulation in Treg cells. In contrast, TCR stimulation downregulated the expression of other mitochondrial proteins (glutamate dehydrogenase 1 and 2 [GLUD1 and 2], mitochondrial ATP synthase subunit β [ATP5B], and mitochondrial HSPA9 in the cytoplasm) (Figure 2B and Table S2C). Also, TCR stimulation in Treg cells increased the expression of ribosomal proteins in the membrane compartment, suggesting an induction of a high biosynthetic profile little impacting glycolysis.
Figure 2.
Proteomic and Metabolic Profiles of In Vitro Anergic and Proliferating Human Treg Cells
Maps of the interactome networks controlling the main biological processes/functional classes associated to metabolism, obtained by comparing unstimulated Treg cells with in vitro cultured Treg cells stimulated with anti-CD3 and anti-CD28 in the membranes (A) and in the cytosol (B), and by comparing anti-CD3 and anti-CD28-stimulated Treg cells with leptin-neutralized Treg cells, in the membranes (C) and in the cytosol (D). Red plots correspond to specific proteins upregulated in anti-CD3 and anti-CD28-stimulated Treg cells; blue plots correspond to proteins downregulated in anti-CD3 and anti-CD28-stimulated Treg cells (A and B) and to proteins upregulated in leptin-neutralized-Treg cells (C) and (D); pink plots represent the equally distributed proteins in the two experimental conditions; white plots are the not-identified proteins.
For lipid-related metabolism, our analysis revealed that a key enzyme of FAO, HADH (Figure 2B and Table S2C) was downregulated by TCR stimulation, whereas other proteins belonging to the same functional class were upregulated by TCR stimulation (3-hydroxyacyl-CoA dehydrogenase type-2 [HSD17B10], delta(3,5)-delta(2,4)-dienoyl-CoA isomerase [ECH1], α-1-antichymotrypsin [Serpin A3]) (Figure 2A and Table S2C). Moreover, a decreased expression of several apolipoproteins (ApoE, ApoC-3, ApoA4) was observed after TCR stimulation (Figures 2A and 2B and Table S2C).
We have previously shown that neutralization of the hormone-cytokine leptin during activation of Treg cells reverses their anergic state and sustains their own proliferation (De Rosa et al., 2007). Here we compared the proteomic profile of in vitro anergic versus proliferating Treg cells (Tables S3A–S3C and Figures S3A and S3B). Treg cell proliferation (induced by leptin neutralization) associated with an increased expression of glycolytic enzymes both in the membrane (2-oxoglutarate dehydrogenase, L-lactate dehydrogenase B chain, LDHB) and, more markedly, in the cytoplasm (L-lactate dehydrogenase α [LDHA] and β chain [LDHB], PGAM1, TALDO1, triosephosphate isomerase [TPI1]) (Figures 2C and 2D and Table S3C). Moreover, several ribosomal proteins were induced by leptin neutralization, in accordance with increased protein synthesis in actively proliferating cells. Moreover, proliferating Treg cells upregulated the expression of several apolipoproteins (ApoE, ApoA4, ApoA2) and proteins associated with FAO (3-hydroxyacyl-CoA dehydrogenase type-2 [HSD17B10], Δ(3,5)-Δ(2,4)-dienoyl-CoA isomerase, mitochondrial [ECH1], HADH) in the membrane and cytosol (Figures 2C and 2D and Table S3C). In contrast, proteins belonging to this functional class (ApoA1, ApoA4, clusterin [CLU], ApoA2, ApoC1, α-1-antichymotrypsin) were downregulated in the cytoplasm (Figure 2D and Table S3C). With regard to mitochondrial proteins, proliferating Treg cells increased the expression of mitochondrial respiratory chain-associated proteins (NADH dehydrogenase α subcomplex 9 [NDUFA9], cytochrome b-c1 complex subunit 1 [UQCRC1], SLC25A3) in the membranes and TCA proteins (MDH2) and ATP synthase subunit β (ATP5B) in the cytoplasm, indicating that Treg cells increased mitochondrial activity to sustain their own proliferation.
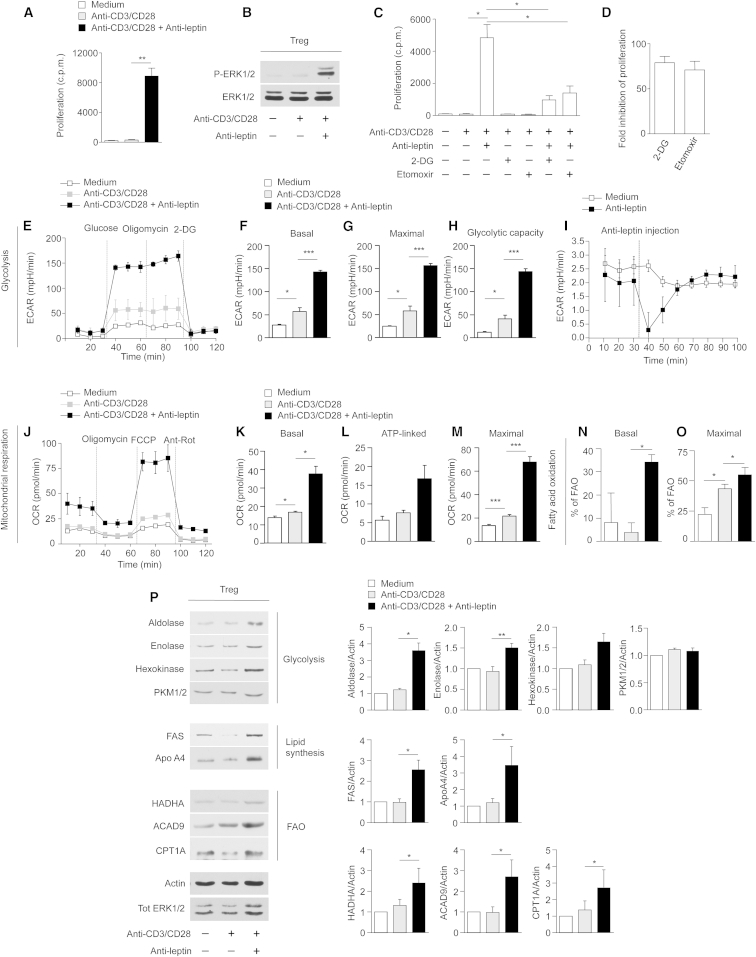
Human Treg Cell Proliferation In Vitro Requires Both Glycolysis and Fatty-Acid Oxidation
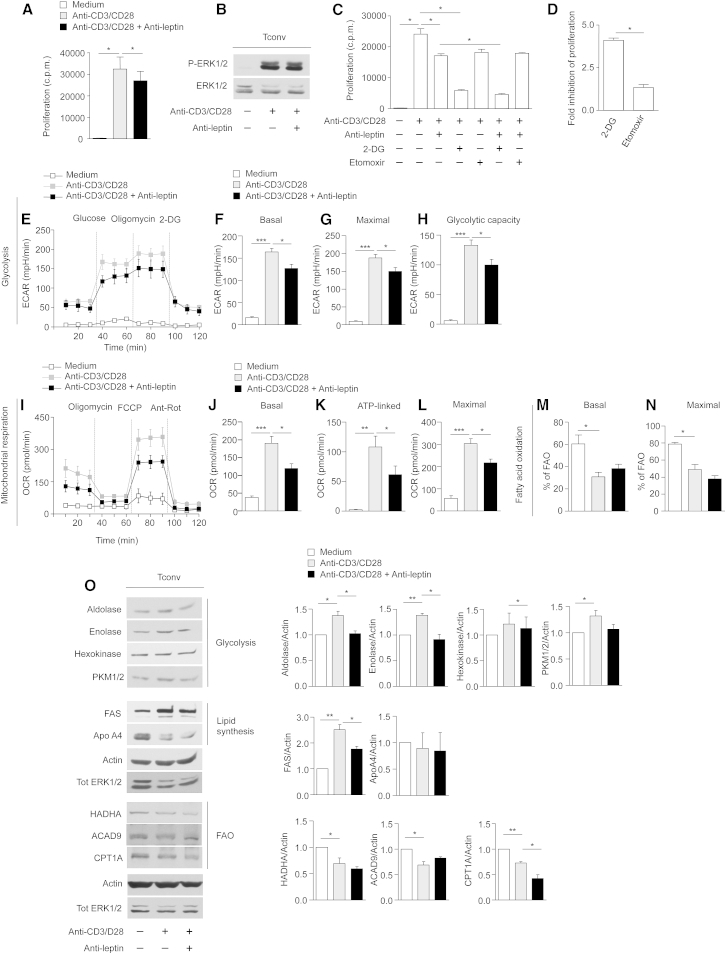
Leptin neutralization in anti-CD3 and anti-CD28-activated Treg cells reversed their anergy (Figure 3A), as indicated by in vitro proliferation and induction of the extracellular signal-regulated kinases 1 and 2 (ERK1/2) phosphorylation (Figure 3B). To evaluate the impact of glycolysis and FAO on Treg cell proliferation in vitro, cells were cultured in the presence or in the absence of specific inhibitors of glycolysis (2-deoxy-D-glucose, 2-DG) and FAO (etomoxir) (Figure 3C). Addition of 2-DG and etomoxir to proliferating Treg cells inhibited their expansion similarly, as testified by the decreased 3H thymidine and bromodeoxyuridine (BrdU) incorporation, thus suggesting the engagement of both pathways in Treg cell proliferation in vitro (Figures 3C and 3D and Figure S4A). Moreover, 2-DG and etomoxir treatment also downregulated the expression of CD25 (in terms of mean fluorescence intensity, MFI) in proliferating Treg cells (Figure S4B). Reduced activation/proliferation of Treg cells, secondary to the metabolic perturbations, were not associated with either altered Treg cells survival or viability, as indicated by propidium iodide (PI) and Annexin V (AV) staining (Figures S4C and S4D). We also analyzed the bioenergetic profile of these cells evaluating ECAR and OCR. Anergic (both unstimulated and TCR-stimulated) Treg cells did not properly respond either to glucose, oligomycin, or 2-DG treatment (Figure 3E). On the contrary, proliferating Treg cells (induced by leptin neutralization) promptly engaged glycolysis, as testified by a sharp increase in ECAR levels evaluated as basal, maximal ECAR (oligomycin-stimulated) and glycolytic capacity (Figures 3E–3H); these data were in agreement with their proteomic profile.
Figure 3.
Human Treg Cell Proliferation In Vitro Requires Both Glycolysis and FAO
(A) In vitro proliferation of Treg cells upon 72 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody. The data are shown as mean ± SEM (n = 6).
(B) Immunoblot for P-ERK1/2 on human Treg cells upon 12 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody. Total ERK1/2 served as a loading control. One representative out of three independent experiments is shown.
(C) In vitro proliferation of Treg cells upon 72 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody, 2-DG, or Etomoxir, alone or in combination. The data are shown as mean ± SEM (n = 3).
(D) Fold inhibition of Treg cell proliferation upon leptin neutralization in the presence of 2-DG or Etomoxir. The data are shown as mean ± SEM (n = 3).
(E) Kinetic profile of ECAR in Treg cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). The data are shown as mean ± SEM of duplicates. ECAR was measured in real time, under basal conditions and in response to glucose, oligomycin and 2-DG. Indices of glycolytic pathway activation, calculated from Treg cells ECAR profile: basal ECAR (F), maximal ECAR (G), and glycolytic capacity (H) in Treg cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). Data are expressed as mean ± SEM of three measurements, each of them in duplicates.
(I) ECAR profile of unstimulated Treg cells before and after leptin-neutralizing antibody injection (one representative out of three independent experiments). The data are shown as mean ± SEM of triplicates.
(J) Kinetic profile of OCR in Treg cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). The data are shown as mean ± SEM of duplicates. OCR was measured in real time, under basal conditions and in response to indicated mitochondrial inhibitors: oligomycin, FCCP, Antimycin A and Rotenone. Indices of mitochondrial respiratory function, calculated from Treg cells OCR profile: basal OCR (K), ATP-linked OCR (L), and maximal OCR (M) in Treg cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). Data are expressed as mean ± SEM of three measurements, each of them in duplicates.
(N and O) Percentage of FAO in Treg cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr in the presence or absence of leptin-neutralizing antibody. FAO was evaluated in basal conditions (N) and during maximal respiration (O) (one representative out of three independent experiments). Data are expressed as mean ± SEM of three measurements, each of them in duplicates.
(P) Immunoblot for aldolase, enolase, hexokinase, and PKM1/2, FAS, ApoA4, HADHA, ACAD9, CPT1A, on Treg cells upon 12 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody. Actin and total ERK 1/2 served as a loading control. One representative out of two independent experiments is shown. The graphs show the relative densitometric quantitation of aldolase, enolase, hexokinase, PKM1/2, FAS, ApoA4, HADHA, ACAD9, and CPT1A normalized on actin in unstimulated (white columns), anti-CD3, and anti-CD28-stimulated (gray columns) and leptin-neutralized Treg cells (black columns) and shown as fold over unstimulated Treg cells (n = 6; data are shown as mean ± SEM of at least two independent experiments, in triplicates). All statistical analysis by paired two-tailed Student’s t test. (∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001).
Because Treg cells proliferation had been obtained through metabolic manipulation (leptin neutralization), we also evaluated whether acute leptin neutralization during ECAR measurement (Seahorse assays) could affect its profile. Specifically, we found that ECAR levels were reduced by acute leptin neutralization before TCR engagement (Figure 3I). These data suggest that the inhibition of glycolysis before TCR stimulation (by leptin neutralization) was necessary to render Treg cells responsive to proliferative stimuli allowing the reversal of their anergic state (Figure 3I).
Also, to assess whether leptin neutralization could influence mitochondrial functions, we measured OCR in basal conditions, after addition of oligomycin, carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), and antimycin-A/rotenone (to inhibit electron transport chain, ETC) (Figure 3J). OCR was induced following leptin neutralization, as indicated by an increased basal, ATP-linked, and maximal respiratory capacity in proliferating Treg cells, whereas all these parameters were lower in unstimulated and TCR-stimulated Treg cells (Figures 3K–3M). Moreover, proliferating Treg cells had increased FAO as compared to anergic (unstimulated and TCR-stimulated) Treg cells, both in basal and maximal respiration conditions (Figures 3N and 3O and S4E and S4F). Biochemical analysis confirmed these data, showing an increased expression of aldolase, enolase, and hexokinase (glycolytic enzymes), FAS, ApoA4 (lipid metabolism-related proteins), and HADHA, ACAD9, carnitine palmitoyltransferase 1A (CPT1A) (FAO enzymes) in proliferating Treg cells as compared to anergic (both unstimulated and TCR-stimulated) Treg cells (Figures 3P and S4G–S4I). Moreover, we found that treatment with 2-DG or etomoxir in proliferating Treg cells partially blunted the changes in the amount of aldolase (Figures S4J and S4K) and HADHA (a FAO protein) (Figures S4L and S4M), induced by leptin neutralization; this evidence confirmed specificity of the phenomena.
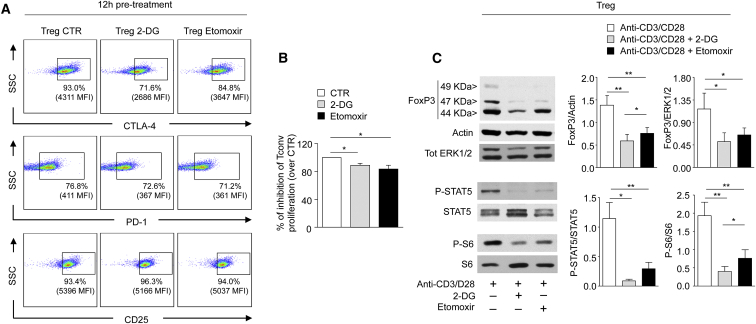
Inhibition of Glycolysis and FAO Alters the Suppressive Function of Human Treg Cells
To determine whether metabolic perturbations could associate with a change in the regulatory function of human Treg cells, we analyzed the expression of several Treg-lineage associated markers (i.e., CD25, PD-1, and CTLA4) in unstimulated Treg cells upon 12 hr pre-treatment with either 2-DG or etomoxir. The expression of CTLA-4 and PD-1 was down-modulated by the above treatments (Figure 4A), as compared to untreated Treg cells (CTR), whereas slight differences were observed in CD25 expression (in terms of MFI). Next, Treg cells pre-treated with 2-DG or etomoxir were tested for their capability to suppress in vitro CFSE-labeled Tconv cells proliferation in co-colture experiments. Inhibition of glycolysis or FAO resulted in a significant reduction (about 20%) of Treg cell suppressive function, as compared to control Treg cells (Figure 4B).
Figure 4.
Suppressive Function of Human Treg Cells Requires Both Glycolysis and FAO
(A) Representative flow cytometry plots showing expression of lineage specific markers (CTLA-4, PD-1, CD25) in Treg cells upon 12 hr pretreatment with vehicle (CTR), 2-DG, or etomoxir. Percentage and MFI of positive cells are indicated. One representative out of three independent experiments is shown.
(B) Percentage of inhibition of CFSE-labeled Tconv cell proliferation co-cultured for 72 hr with Treg cells pre-treated 12 hr with vehicle, 2-DG, or etomoxir. The data are shown as mean ± SEM and are expressed as reduction over CTR (n = 8).
(C) Immunoblot for FoxP3, actin, total ERK1/2, P-STAT5, STAT5, P-S6, and S6 on Treg cells upon 12 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of 2-DG or etomoxir. One representative out of two independent experiments is shown. The graphs show the relative densitometric quantitation of FoxP3 (normalized on actin and total ERK 1/2), P-STAT5 (normalized on total STAT5), and P-S6 (normalized on total S6) (n = 6, data are shown as mean ± SEM of two independent experiments, in triplicates). All statistical analysis by paired two-tailed Student’s t test (∗p < 0.05, ∗∗p < 0.005).
To better understand the molecular mechanisms of the above findings, we studied the effects of either 2-DG or etomoxir on FoxP3 expression and cytokine-related- and cell metabolism-associated signaling in Treg cells. FoxP3 expression was reduced in Treg cells treated with 2-DG or etomoxir, as compared to control Treg cells (Figure 4C). Because the IL-2-IL-2R signaling pathway has a pivotal role in the generation, in vivo proliferation, and function of Treg cells (Tang et al., 2008, Long et al., 2010, Cheng et al., 2011, Cheng et al., 2013), we evaluated Signal Transducer and Activator of Transcription-5 (STAT-5) phosphorylation as the major element of transduction for IL-2R. 2-DG and etomoxir markedly reduced STAT-5 phosphorylation in Treg cells stimulated for 12 hr with anti-CD3 and anti-CD28 (Figure 4C). This phenomenon was accompanied by a reduced phosphorylation of the ribosomal protein S6, a downstream target of mTOR, whose activity is pivotal for Treg cell biology (Zeng et al., 2013, Procaccini et al., 2010) (Figure 4C).
Proteomic and Metabolic Profiles of In Vitro Cultured Human Tconv Cells
The proteomic profile of Tconv cells, in basal conditions and after TCR stimulation or metabolic manipulation, through leptin neutralization, was also evaluated (Tables S4 and S5 and Figures S5 and S6). Glycolytic enzymes (ENO1, TPI1, ALDOA, PGMA1, and PGK2) were strongly upregulated by TCR stimulation, particularly in the cytoplasm (Figure 5B and S5B and Tables S4B and S4C) and the same occurred for apolipoproteins (ApoE, ApoA4, ApoC3). In the membrane compartment, TCR stimulation in Tconv cells induced the expression of several ribosomal proteins, in agreement with induction of protein synthesis and proliferation (Figures 5A and S5A and Tables S4A and S4C). This phenomenon was associated with a reduction of several mitochondrial proteins (VDAC3, ATP5F1, ATP5H), namely those connected to the Krebs cycle (CS, SDHA) and mitochondrial permeability transition (SLC25A3), together with a reduction of FAO enzyme (HADH) (Figures 5A and S5A and Tables S4A and S4C).
Figure 5.
Proteomic and metabolic profiles of in vitro-cultured human Tconv cells
Maps of the interactome networks controlling the main biological processes/functional classes associated to metabolism, obtained by comparing unstimulated Tconv cells with in vitro cultured Tconv cells stimulated with anti-CD3 and anti-CD28 in the membranes (A) and in the cytosol (B), and by comparing anti-CD3 and anti-CD28-stimulated Tconv cells with leptin-neutralized Tconv cells, in the membranes (C) and in the cytosol (D). Red plots correspond to specific proteins upregulated in anti-CD3 and anti-CD28-stimulated Tconv cells; blue plots correspond to proteins downregulated in anti-CD3 and anti-CD28-stimulated Tconv cells (A) and (B) and to proteins upregulated in leptin-neutralized Tconv cells (C) and (D); pink plots represent the equally distributed proteins in the two experimental conditions; white plots are the not-identified proteins.
We have reported that metabolic manipulation, through leptin neutralization had opposite effects on Treg and Tconv cells, because it induces Treg cell proliferation while it inhibits Tconv cell proliferation (De Rosa et al., 2007). We found that leptin neutralization of Tconv cells associated with a consistent reduction of ribosomal proteins in membranes, as well as a decreased expression of mitochondrial proteins associated with electron transport chain (SDHA, ATP synthase beta chain [ATP5B], VDAC1 and VDAC3) or TCA cycle (MDH2), both in the membrane and in the cytoplasm (Figures 5C and 5D and S6A and S6B and Tables S5A–S5C). Inhibition of Tconv cell proliferation by leptin neutralization reduced a fraction of glycolytic enzymes such as ALDOA and PGK1 and 2 but upregulated other intermediates such as LDHB, TPI1, GAPDH, and ENO3 (Figures 5C and 5D and S6A and S6B and Tables S5A–S5C).
With regard to lipid metabolism, in the membrane compartment leptin neutralization downregulated the expression of FAO-associated proteins (hydroxysteroid [17-beta] dehydrogenase 4 [HSD17B4], HADHA) and apolipoproteins (ApoB), but upregulated the expression of other proteins of the same functional class (annexin A I [ANXA1], mitochondrial short-chain enoyl-CoA hydratase [ECHS1], ApoA1, ApoA4, ApoA2) (Figures 5C and S6A and Tables S5A and S5C). In the cytoplasm, leptin neutralization downregulated the expression of ApoC3 and ApoA4 and increased the expression of ApoA2, Serpin A3, and leukotriene A4 hydrolase (LTA4H) (Figures 5D and S6B and Tables S5B and S5C).
Human Tconv Cell Proliferation In Vitro Requires Glycolysis but Not Fatty-Acid Oxidation
TCR stimulation of Tconv cells induced their proliferation associated with the phosphorylation of the ERK1/2 kinases (Figures 6A and 6B), whereas leptin neutralization was able to inhibit their proliferation. Culture of Tconv cells in the presence or in the absence of 2-DG and etomoxir showed that the inhibition of glycolysis strongly reduced Tconv cell proliferation (evaluated by 3H thymidine incorporation and BrdU staining) (Figures 6C and 6D and S7A), also in terms of CD25 expression (Figure S7B), without affecting their viability (Figures S7C and S7D), with an additive effect on inhibition mediated by leptin neutralization. Conversely, inhibition of FAO with etomoxir did not affect Tconv cell proliferation and survival, suggesting that these cells preferentially use glycolysis to sustain their own proliferation (Figures 6C and 6D and Figures S7A–S7D).
Figure 6.
Human Tconv Cell Proliferation In Vitro Requires Glycolysis but Not FAO
(A) In vitro proliferation of Tconv cells upon 72 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody. The data are shown as mean ± SEM (n = 6).
(B) Immunoblot for P-ERK1/2 on human Tconv cells upon 12 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody. Total ERK1/2 served as a loading control. One representative out of three independent experiments is shown.
(C) In vitro proliferation of Tconv cells upon 72 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody, 2-DG, or etomoxir, alone or in combination. The data are shown as mean ± SEM (n = 3).
(D) Fold inhibition in Tconv cells proliferation upon TCR-mediated stimulation in the presence of 2-DG and etomoxir. The data are shown as mean ± SEM (n = 3).
(E) Kinetic profile of ECAR in Tconv cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). The data are shown as mean ± SEM of triplicates. ECAR was measured in real time, under basal conditions and in response to glucose, oligomycin, and 2-DG. Indices of glycolytic pathway activation, calculated from Tconv ECAR profile: basal ECAR (F), maximal ECAR (G), and glycolytic capacity (H) in Tconv cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). Data are expressed as mean ± SEM of three measurements, each of them in triplicates.
(I) Kinetic profile of OCR in Tconv cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). The data are shown as mean ± SEM of triplicates. OCR was measured in real time, under basal conditions and in response to indicated mitochondrial inhibitors: oligomycin, FCCP, Antimycin A, and Rotenone. Indices of mitochondrial respiratory function, calculated from Tconv OCR profile: basal OCR (J), ATP-linked OCR (K), maximal OCR (L) in Tconv cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr, in the presence or absence of leptin-neutralizing antibody (one representative out of three independent experiments). Data are expressed as mean ± SEM of three measurements, each of them in triplicates.
(M and N) Percentage of FAO in Tconv cells stimulated or not with anti-CD3 and anti-CD28 for 12 hr in the presence or absence of leptin-neutralizing antibody. FAO was evaluated in basal conditions (M) and during maximal respiration (N) (one representative out of three independent experiments). Data are expressed as mean ± SEM of three measurements, each of them in duplicates.
(O) Immunoblot for Aldolase, Enolase, Hexokinase, and PKM1/2, Fatty acid synthase (FAS), ApoA4, HADHA, ACAD9, CPT1A on Tconv cells upon 12 hr anti-CD3, and anti-CD28 stimulation in the presence or absence of leptin-neutralizing antibody. Actin and total ERK 1/2 served as a loading control. One representative out of three independent experiments is shown. The graphs show the relative densitometric quantitation of Aldolase, Enolase, Hexokinase, PKM1/2, FAS, ApoA4, HADHA, ACAD9, and CPT1A normalized on actin in unstimulated (white columns), anti-CD3 and anti-CD28-stimulated (gray columns), and leptin-neutralized Tconv cells (black columms) and shown as fold over unstimulated Tconv cells. (n = 6; data are shown as mean ± SEM of two independent experiments, in triplicates). All statistical analysis by paired two-tailed Student’s t test. (∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001).
Confirming the above data, TCR stimulation induced an increase in basal, maximal ECAR, and glycolytic capacity in Tconv cells, whereas leptin neutralization inhibited these parameters (Figures 6E–6H). Interestingly, basal, ATP-linked, and maximal OCR were upregulated during Tconv cell activation but leptin neutralization inhibited their TCR-mediated induction (Figures 6I–6L). Concomitantly, Tconv cell activation with anti-CD3 and anti-CD28 associated with a strong reduction of FAO as compared to unstimulated Tconv cells. Leptin neutralization had no effect on the modulation of this pathway, suggesting that Tconv cells did not use FAO to sustain ATP production and proliferation, but rather switched T cell metabolism from FAO to aerobic glycolysis (Figures 6M and 6N and S7E and S7F).
Finally, we confirmed the above data at the biochemical level by Western blotting (Figures 6O and S7G–S7I). TCR stimulation induced aldolase, enolase, and PKM1/2 (glycolytic enzymes) and FAS, whereas ACAD9, CPT1A, and HADHA (all involved in FAO) were downregulated by TCR-mediated activation. In agreement with the metabolic data, leptin neutralization reduced the expression of glycolytic enzymes but had little effect on FAO modulation (Figures 6O and S7G–S7I). Of note, the inhibition of glycolytic and FAO proteins induced by leptin neutralization has been confirmed by treatment with 2-DG or etomoxir, which equally inhibited their expression (Figures S7J–S7M).
Effector Functions of Human Tconv Cells Are Mainly Sustained by Glycolysis
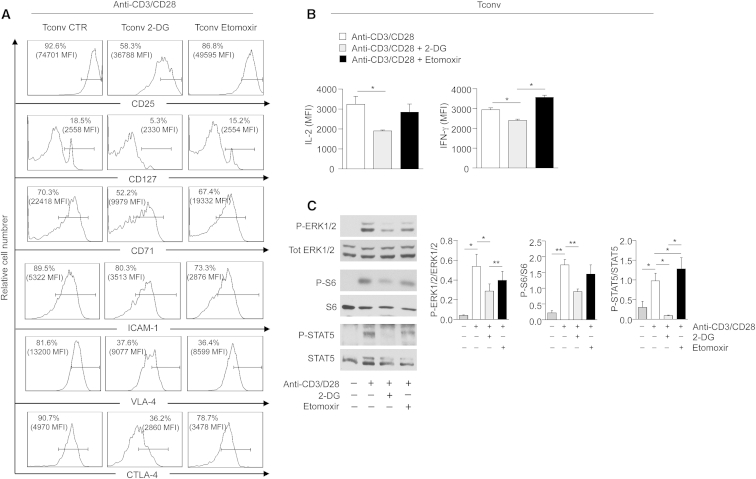
To study the effects of either glycolysis or FAO inhibition on Tconv cell function, we evaluated the expression of several activation markers (i.e., CD25, CD127, CD71, ICAM-1, VLA-4, and CTLA-4) on Tconv cells upon TCR stimulation, in the presence or the absence of etomoxir or 2DG, respectively (Figure 7A). As for proliferation, 2-DG treatment reduced expression of all these markers while etomoxir only affected expression of ICAM-1 and VLA-4, suggesting that Tconv cell activation mainly relies on glycolysis. Consistent with the reduced proliferation and activation of Tconv cells, inhibition of glycolysis associated with a decreased production of pro-inflammatory cytokines, such as IL-2 and IFN-γ, whereas etomoxir had a little impact on the secretion of these cytokines by Tconv cells (Figure 7B).
Figure 7.
Effector Functions of Human Tconv Cells Are Mainly Sustained by Glycolysis
(A) Representative flow cytometry plots showing expression of activation markers (CD25, CD127, CD71, ICAM-1, VLA-4, and CTLA-4) in Tconv cells upon 36 hr activation with anti-CD3 and anti-CD28 in the presence or absence of 2-DG or etomoxir. Percentage and MFI of positive cells are indicated. One representative out of two independent experiments is shown.
(B) MFI of IL-2 and IFN-γ expression in Tconv cells stimulated for 12 hr with anti-CD3 and anti-CD28 in the presence or absence of 2-DG or etomoxir, analyzed by flow cytometry. Data are expressed as mean ± SEM (n = 3).
(C) Immunoblot for P-ERK 1/2, P-STAT5, and P-S6 on Tconv cells upon 12 hr anti-CD3 and anti-CD28 stimulation in the presence or absence of 2-DG or etomoxir. Total ERK 1/2, total STAT5, and total S6 served as a loading control. One representative out of two independent experiments is shown. The graphs show the relative densitometric quantitation of each phosphorylated protein normalized on its total form, in Tconv cells treated for 12 hr with anti-CD3 and anti-CD28 in the presence or absence of 2-DG or etomoxir (n = 6, data are shown as mean ± SEM of two independent experiments, in triplicates). All statistical analysis by paired two-tailed Student’s t test (∗p < 0.05, ∗∗p < 0.005).
We next evaluated the activity of the main biochemical pathways related to T cell cycle/proliferation, IL-2-IL-2R signaling and mTOR pathway in Tconv cells, upon TCR-mediated stimulation in the presence or the absence of etomoxir or 2-DG (Figure 7C). As for the previous parameters, 2-DG treatment significantly reduced the phosphorylation of ERK1/2, S6, and STAT5, when compared to anti-CD3 and anti-CD28-activated Tconv cells, whereas etomoxir did not affect significantly their phosphorylation (Figure 7C).
Finally, in order to dissect the effects of glycolysis and FAO on Tconv cell metabolism, we also performed Seahorse experiments in Tconv cells chronically treated with 2-DG or etomoxir. ECAR profile of activated Tconv cells was not affected by 12 hr treatment with etomoxir, differently from what observed with 2-DG (Figures S7N–S7Q). At the same time, FAO rate of activated/proliferating Tconv cells was comparable to that of etomoxir-treated Tconv cells, in which FAO was chronically inhibited. On the contrary, inhibition of glycolysis with 2-DG enhanced FAO. Taken together, these data suggest that FAO only marginally participates to Tconv cell activation/proliferation (Figures S7R–S7U), as they are more sensitive to inhibition of glycolysis, which represents the main metabolic pathway in the control of their function.
Discussion
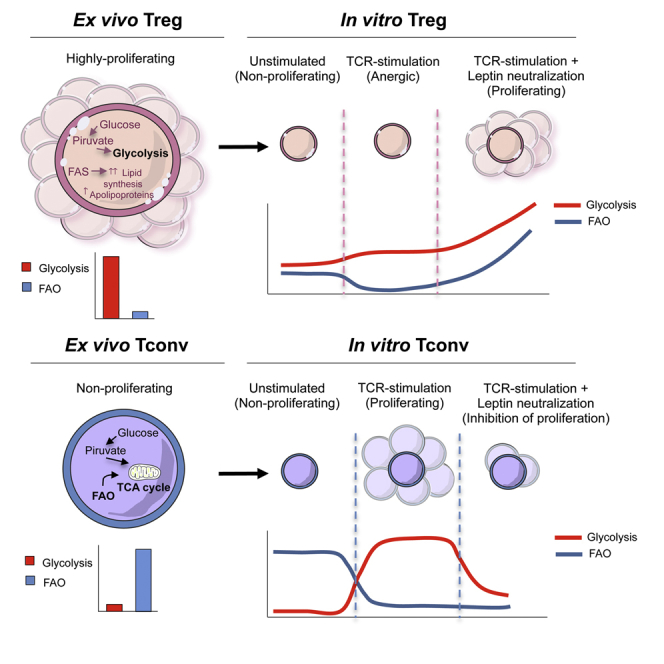
Understanding the impact of basic metabolic pathways on the development and function of immune cells is of great interest in immunological research. Our study illustrates a proteomic and metabolic comparison between human Treg and Tconv cells in both unmanipulated (freshly-isolated) and in vitro culture conditions. This study identified (1) ex vivo metabolic programs associated with cell lineage commitment and diversification; (2) the differential impact of metabolism on the proteomic network and its dynamic changes occurring in in vitro cultured Treg versus Tconv cells; (3) the impact of metabolism on Treg and Tconv cell function; and (4) metabolic regulation of human Treg cell homeostasis. Our findings unveil the metabolic signature of ex vivo human Treg and Tconv cells and provide evidence of how these cells can adapt their metabolic requirements according to the in vitro microenvironmental cues to support their specific functions. This might have major implications in understanding the impact of metabolism on immune tolerance, both in physiological conditions and in autoimmunity.
To identify the proteomic networks associated with the biology of human Treg and Tconv cells, we performed proteomic profiling in unmanipulated, freshly-isolated cells. We suggest that the main changes in protein profiles between human Treg and Tconv cells can be attributed to a different fitness in their metabolic asset (MacIver et al., 2013). Michaleck and colleagues showed that although Th1 and Th17 cells (which express high amounts of the glucose transporter Glut1) are highly glycolytic, Treg cells generated through in vitro polarization of CD4+ T cells preferentially use lipid oxidation (Michalek et al., 2011, Gerriets et al., 2015). Because those differentiating conditions were induced in vitro by addition of high doses of cytokines (TGF-β), we investigated the metabolism associated with unmanipulated, freshly-isolated human Treg and Tconv cells. We found that unmanipulated human Treg cells were highly glycolytic at the proteomic, biochemical, and functional level as compared to Tconv cells. Because glycolysis is believed to promote biosynthesis for rapid T cell growth, this result is in agreement with previous finding showing that Treg cells actively proliferate in vivo both in humans and in mice (Vukmanovic-Stejic et al., 2006, Vukmanovic-Stejic et al., 2008) and are also metabolically more active than Tconv cells, expressing higher level of mammalian-target of rapamycin (mTOR) (Procaccini et al., 2010)—whose activity associates with high glycolytic levels (Tandon et al., 2011, Cheng et al., 2014). Glycolysis is important for generation of inducible Treg (iTreg) cells via the induction of specific Foxp3 splicing variants (De Rosa et al., 2015). In this context, human Treg cells can acidify the medium, even in the absence of exogenous glucose, because of the high lactate production following proliferation in vivo; as consequence of this state, upon injection of glucose, glycolysis is further increased in Treg cells, reaching levels which were always higher than Tconv cells.
Our finding is in line with previous data that showed that Treg cells produce high amounts of leptin and express high level of leptin receptor (LepR) (De Rosa et al., 2007). LepR activation induces glucose uptake and license metabolically CD4+ T cells for proliferation. We also found that freshly-isolated human Treg cells displayed higher amount of apolipoprotein when compared with Tconv cells, in agreement with the finding that T cell proliferation also requires glycolytic-lipogenic metabolic pathways (Berod et al., 2014). These findings resemble the metabolic asset of activated CD4+ T cells, which rely on glycolysis for de novo fatty-acids synthesis to support the need for membrane synthesis required for proliferation. In this context, the high amount of apoA-IV proteins expressed by Treg cells further supports the anti-inflammatory and anti-atherogenic role of apolipoproteins (Vowinkel et al., 2004, Bellemore et al., 2014, Nilsson et al., 2012, Wilhelm et al., 2010). Finally, we observed higher mitochondrial respiration in freshly-isolated Treg cells, possibly associated with their enhanced capability to respond to metabolic demand. Instead, freshly-isolated Tconv cells expressed high levels of TCA cycle-associated proteins and used FAO to sustain proliferation, reinforcing the notion that resting Tconv cells maintain low rates of glycolysis and predominantly use FAO to produce ATP (MacIver et al., 2013, Maciolek et al., 2014).
Treg cell biology is strictly linked to adipose tissue homeostasis. In this context leptin, an adipose-tissue-derived cytokine, constrains Treg cell proliferation by activating the mTOR pathway and inducing TCR hyporesponsiveness (De Rosa et al., 2007). Leptin neutralization reversed anergy and induced Treg cell proliferation while inhibiting Tconv proliferation (De Rosa et al., 2007). Because of those data, we used leptin neutralization for in vitro expansion of Treg cells concomitantly to inhibit Tconv proliferation. This approach allowed us to analyze the metabolic changes of Treg and Tconv cells in different culture conditions and TCR stimulation. We found that anergic Treg cells expressed high amounts of ribosomal proteins, suggesting an active biosynthetic activity accompanied by the maintenance of a high glycolytic rate (also supported by high mTOR activity) not associated with FAO. In contrast, in vitro Treg cell proliferation, induced by leptin neutralization, required activation of both glycolysis and FAO. At the functional level, we observed that glycolysis and FAO inhibition downregulated the expression of FoxP3 and other specific Treg cell markers, reducing their suppressive activity. At the molecular level, these events associated with functional impairment of IL-2-IL-2R-STAT5 signaling pathway, in line with reports showing that IL-2 signaling is essential for functional maturation of Treg cells, through activation of STAT5 (Cheng et al., 2011, Cheng et al., 2013, Burchill et al., 2007), which is a critical factor in the induction and maintenance of FoxP3 expression (Passerini et al., 2008, Carbone et al., 2014).
The mTOR pathway controls TCR-dependent FoxP3 expression (Sauer et al., 2008). The anergic state and suppressive function of Treg cells in vitro partly depended on their high metabolic rate, associated with mTOR pathway activation. Here we observed that reduced Treg cell suppressive capacity, induced by 2-DG and etomoxir, associated with an impaired mTOR activity, thus suggesting that the metabolic control is crucial for Treg cell functions. Of note, Tconv cells had an opposite behavior, because they preferentially used glycolysis to sustain their proliferation, while reducing FAO, in line with recent findings showing that Th1 and Th17 pathogenic cells require glycolysis to allow full differentiation (Gerriets et al., 2015). Functionally, inhibition of glycolysis impaired Tconv cell activation by downmodulating ERK1 and ERK2 and STAT5 and S6 phosphorylation and modulating effector functions through reduction of pro-inflammatory cytokines.
Taken together, our data suggest that Treg cells are highly sensitive to metabolic perturbations as compared to Tconv cells. Glycolysis in Treg cells made them unresponsive to proliferative stimuli in vitro. A transient inhibition of glycolysis reversed Treg cell anergy, restoring responsiveness to TCR activation; this is in agreement with the finding that transient mTOR inhibition could reverse Treg cell anergy and with the work showing that knocking down Glut1 increased Treg cell expansion in vivo (Macintyre et al., 2014).
To summarize, this study shows that (1) Treg cells are very sensitive to metabolic demand and are highly glycolytic ex vivo; (2) Treg cell anergy in vitro is associated with high glycolysis, since its transient inhibition (through leptin neutralization) reverses this state; and (3) in vitro proliferation and function of Treg cells require both glycolysis and FAO, whereas those of Tconv cells require only glycolysis. The discrepancy between ex vivo and in vitro data raised the question of whether in vitro metabolic manipulations (i.e., additions of high doses of cytokines) might have a possible impact on the way in which human Treg cells are usually expanded in vitro for possible therapeutic potential. A recent report showed that in a phase I study, the adoptive transfer of autologous ex vivo-expanded human Treg cells from type I diabetes patients had no therapeutic effect on their metabolic dysfunction and C-peptide production, insulin use and hemoglobin A1c (HbA1c) levels (Bluestone et al., 2015). It was suggested that the progressive decline in circulating Treg cells after transfer could be ascribed to cell death linked to reduced IL-2 signaling associated with the shift from in vitro to in vivo (Bluestone et al., 2015). Our data suggest engagement of different metabolic pathways during the expansion of Treg cells in vivo versus in vitro and different metabolic and proteomic profiles under the two conditions. As such, our report identifies potential metabolic regulators of Treg and Tconv cell proliferation and responsiveness to stimulation that could unveil networks relevant for the control of T cell homeostasis and function.
Experimental Procedures
Cell Purification, Cultures, and Proliferation Assays
The study was approved by the Institutional Review Board (IRB) of the Università degli Studi di Napoli “Federico II.” Buffy coats from control healthy subjects were obtained after they signed an IRB-approved written informed consent. We isolated human Treg (CD4+CD25highCD127−) and Tconv (CD4+CD25−) cells from PBMCs from buffy coats of human healthy donors by high-performance cell sorting (MoFlo, Dako Beckman-Coulter or BD FACS-Jazz, BD Bioscience) in the IEOS-CNR Sorting Facility in Napoli, after staining with the following antibodies at 1:100 dilution: FITC anti-human CD4 (BD PharMingen, clone RPA-T4, Cat.# 555346), PE anti-human CD25 (BD PharMingen, clone M-A251 Cat.# 555432) and APC anti-human CD127 (R&D Systems, clone 40131, Cat.# FAB306A), or by magnetic cell separation with Dynabeads Regulatory T Cell Kit (Invitrogen). Soon after isolation, we rapidly cleaned Treg with Detach reagent (Invitrogen), which removes surface-bound CD25 mAb, and washed them extensively. These CD25-antibody-free Tregs were 95%–98% pure by FACS analysis, and > 95% expressed FoxP3. We cultured cells (2 × 104 cells per well) in round-bottom 96-well plates (Becton Dickinson Falcon) with RPMI 1640 medium supplemented with 100 UI/ml penicillin and 100 μg/ml streptomycin (all from Life Technologies) and 5% AB human serum (Sigma-Aldrich). Cells were stimulated for 3 days in the presence of anti-CD3 and anti-CD28-coated Dynabeads (0.1 beads per cell) (Invitrogen). For in vitro leptin blocking experiments, we used human anti-leptin neutralizing mAb (clone 44802, Cat.# MAB398, R&D Systems) at a final concentration of 20 μg/ml. 2-Deoxy-D-glucose (2DG) (Sigma-Aldrich) was used at final concentration of 1 mM; etomoxir was purchased from Sigma-Aldrich and it was used at the final concentration of 200 μM. On the last day (60–72 hr), we added [3H] thymidine (0.5 μCi per well) (Amersham-Pharmacia Biotech) to the cultures and harvested cells after 12 hr (Tomtec). Radioactivity was measured with a beta-plate scintillation counter (Wallac).
Sample Preparation for Multidimensional Protein Identification Technology Analysis
We stimulated freshly-isolated Tconv and Treg cells either with anti-CD3 and anti-CD28-coated Dynabeads (0.2 beads per cell) (Invitrogen) alone or anti-CD3 and anti-CD28 and anti-leptin-neutralizing mAb for 12 hr. We performed two independent sample preparations for multidimensional protein identification technology (MudPIT) analyses, consisting of 1 × 106 cells per preparation, each deriving from a pool of four independent experiments of Tconv/Treg purifications from different healthy donors. Briefly, total cell lysates were obtained in 50 μl of 50 mM NH4HCO3 and 150 mM NaCl buffer (pH 7.5). Membrane and cytosolic proteins were separated by centrifugation to obtain pellet and supernatant protein samples, respectively. Total protein was quantified using the Bio-Rad protein assay (Bio-Rad).
Tryptic Digestion and Mass Spectrometry
Digestion, detailed mass spectrometry procedures, and data handling for proteomic and network analysis were performed as described in Supplemental Experimental Procedures.
Flow Cytometry, CFSE, and BrdU Staining
For staining of freshly isolated Treg and Tconv and their immunophenotype we used (1:50–100 dilution) Pe-Cy5-labeled anti-CD4 (BD Biosciences, clone RPA-T4, Cat.# 555348), APC-labeled anti-human CD25 (BD Biosciences, clone M-A251, Cat.# 555434), APC anti-human CD71 (BD PharMingen, clone, M-A712 Cat.# 551374), PercP-Cy5.5 anti-human CD127 (BD PharMingen, clone HIL-7R-M21, Cat.# 560551), APC anti-human CD152 (BD PharMingen, clone BNI3 Cat.# 555855), PerCP-Cy5.5 anti-human CD279 (PD-1) (BD PharMingen, clone EH12.1 Cat.# 581273). Thereafter, we washed, fixed and permeabilized cells (fixation-permeabilization buffer, eBioscience) and stained with PE-labeled anti-FoxP3 (eBioscience, clone PCH101, Cat.#12-4776) and FITC-labeled anti-Ki67 (BD Biosciences, clone B56, Cat.# 556026) monoclonal antibodies. Flow cytometry experiments were performed with a BD FACSCanto and analyzed with Diva software from BD and FlowJo software (Tree Star).
Incorporation of Bromodeoxyuridine (BrdU) and FACS analysis were performed with the BrdU Flow Kit (BD-PharMingen) in accordance to the manufacturer’s instructions. The fluorescent dye CFSE (5, 6-carboxyfluorescein diacetate succinimidyl ester) was used at 1 μg/ml (Invitrogen). Flow cytometric analysis of CFSE dilution was performed by gating on CD4+CFSE+ cells stimulated for 96 hr with anti-CD3 and anti-CD28 Dynabeads (0.5 beads/cell) (Invitrogen) alone or co-cultured with Treg cells pre-treated or not with 2-DG or etomoxir for 12 hr.
Mitochondrial Bioenergetics and Metabolic Assays
The metabolic profiles have been evaluated as described in Supplemental Experimental Procedures.
Western Blot Analyses
Total cell lysates and Western blot analysis were performed as previously described (De Rosa et al., 2007). The antibodies used were the following: anti-aldolase, anti-enolase, anti-hexokinase, anti-PKM1/2, anti-FAS, anti-apoA4, anti-HADHA, anti-ACAD9, anti-CPT1A, anti-DLST, anti-SDHA, anti-phospho STAT5, anti-STAT5, anti-phospho-S6, anti-S6 (all 1:1000 dilution and from Cell Signaling Technology), anti-FoxP3 (1:500 dilution from eBioscience). anti-ERK1/2 (1:500 dilution) and anti-phospho-ERK1/2 (1:500 dilution) (from Santa Cruz Biotechnology), The filters were also probed with an actin antibody (1:1000 from Santa Cruz) to normalize for the amount of loaded protein. All filters were quantified as previously described (De Rosa et al., 2007).
Author Contributions
C.P., F.C., P.M., and G. Matarese designed the study; C.P., F.C., D.D.S., F.B., V.D.R., M.G., C.A., D.F., M.C., and A.P. performed the experiments; C.P., F.C., P.M., D.T., G. Marone, and G. Matarese analyzed the data and interpreted the results; C.P. and F.C. performed statistical analyses; C.P., A.L.C., P.M., and G. Matarese wrote the manuscript.
Acknowledgments
G. Matarese is supported by grants from the European Union IDEAS Programme European Research Council Starting Grant “menTORingTregs” n. 310496, the Fondazione Italiana Sclerosi Multipla (FISM) n. 2012/R/11, and the EFSD/JDRF/Lilly Programme 2015. V.D.R. is supported by the Ministero della Salute grant n. GR-2010-2315414, Fondo per gli Investimenti della Ricerca di Base (FIRB) grant n. RBFR12I3UB_004 and the Fondazione Italiana Sclerosi Multipla (FISM) n. 2014/R/21. M.G. is supported by grant from Juvenile Diabetes Research Foundation (JDRF) n. 1-PNF-2015-115-5-B. A.L.C. is partly supported by the National Institutes of Health grant AI109677. P.M. is supported by the Italian Ministry of Economy and Finance to the CNR for the Project “FaReBio di Qualita,” by Italian Ministry of University and Research for the PON projects (01 02388 and 01 01426); by Fondazione Cariplo (2009-2532). The authors wish to thank Maria Rosaria Montagna and Vincenzo Gigantino for technical support and all members of the Laboratory of Immunology at IEOS-CNR for assistance and support. This work is dedicated to the memory of E. Papa and S. Zappacosta.
Published: February 16, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes seven figures, five tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2016.01.028.
Supplemental Information
References
- Bellemore S.M., Nikoopour E., Au B.C., Krougly O., Lee-Chan E., Haeryfar S.M., Singh B. Anti-atherogenic peptide Ep1.B derived from apolipoprotein E induces tolerogenic plasmacytoid dendritic cells. Clin. Exp. Immunol. 2014;177:732–742. doi: 10.1111/cei.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Bluestone J.A., Mackay C.R., O’Shea J.J., Stockinger B. The functional plasticity of T cell subsets. Nat. Rev. Immunol. 2009;9:811–816. doi: 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluestone J.A., Buckner J.H., Fitch M., Gitelman S.E., Gupta S., Hellerstein M.K., Herold K.C., Lares A., Lee M.R., Li K. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci. Transl. Med. 2015;7:315ra189. doi: 10.1126/scitranslmed.aad4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla F., Lavatelli F., Di Silvestre D., Valentini V., Rossi R., Palladini G., Obici L., Verga L., Mauri P., Merlini G. Reliable typing of systemic amyloidoses through proteomic analysis of subcutaneous adipose tissue. Blood. 2012;119:1844–1847. doi: 10.1182/blood-2011-07-365510. [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Carbone F., De Rosa V., Carrieri P.B., Montella S., Bruzzese D., Porcellini A., Procaccini C., La Cava A., Matarese G. Regulatory T cell proliferative potential is impaired in human autoimmune disease. Nat. Med. 2014;20:69–74. doi: 10.1038/nm.3411. [DOI] [PubMed] [Google Scholar]
- Cheng G., Yu A., Malek T.R. T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells. Immunol. Rev. 2011;241:63–76. doi: 10.1111/j.1600-065X.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., Yu A., Dee M.J., Malek T.R. IL-2R signaling is essential for functional maturation of regulatory T cells during thymic development. J. Immunol. 2013;190:1567–1575. doi: 10.4049/jimmunol.1201218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa V., Procaccini C., Calì G., Pirozzi G., Fontana S., Zappacosta S., La Cava A., Matarese G. A key role of leptin in the control of regulatory T cell proliferation. Immunity. 2007;26:241–255. doi: 10.1016/j.immuni.2007.01.011. [DOI] [PubMed] [Google Scholar]
- De Rosa V., Galgani M., Porcellini A., Colamatteo A., Santopaolo M., Zuchegna C., Romano A., De Simone S., Procaccini C., La Rocca C. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 2015;16:1174–1184. doi: 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaie K., von Boehmer H. The impact of CD4+CD25+ Treg on tumor specific CD8+ T cell cytotoxicity and cancer. Semin. Cancer Biol. 2006;16:124–136. doi: 10.1016/j.semcancer.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Kohrt H.E., Pillai A.B., Lowsky R., Strober S. NKT cells, Treg, and their interactions in bone marrow transplantation. Eur. J. Immunol. 2010;40:1862–1869. doi: 10.1002/eji.201040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Godfrey W.R., Porter S.B., Ge Y., June C.H., Blazar B.R., Boussiotis V.A. CD4+CD25+ regulatory T-cell lines from human cord blood have functional and molecular properties of T-cell anergy. Blood. 2005;106:3068–3073. doi: 10.1182/blood-2005-04-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.A., Cerosaletti K., Bollyky P.L., Tatum M., Shilling H., Zhang S., Zhang Z.Y., Pihoker C., Sanda S., Greenbaum C., Buckner J.H. Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4(+)CD25(+) regulatory T-cells of type 1 diabetic subjects. Diabetes. 2010;59:407–415. doi: 10.2337/db09-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.T., Li M.O. Transcriptional control of regulatory T cell development and function. Trends Immunol. 2013;34:531–539. doi: 10.1016/j.it.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre A.N., Gerriets V.A., Nichols A.G., Michalek R.D., Rudolph M.C., Deoliveira D., Anderson S.M., Abel E.D., Chen B.J., Hale L.P., Rathmell J.C. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciolek J.A., Pasternak J.A., Wilson H.L. Metabolism of activated T lymphocytes. Curr. Opin. Immunol. 2014;27:60–74. doi: 10.1016/j.coi.2014.01.006. [DOI] [PubMed] [Google Scholar]
- MacIver N.J., Michalek R.D., Rathmell J.C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri P., Scarpa A., Nascimbeni A.C., Benazzi L., Parmagnani E., Mafficini A., Della Peruta M., Bassi C., Miyazaki K., Sorio C. Identification of proteins released by pancreatic cancer cells by multidimensional protein identification technology: a strategy for identification of novel cancer markers. FASEB J. 2005;19:1125–1127. doi: 10.1096/fj.04-3000fje. [DOI] [PubMed] [Google Scholar]
- Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., Rathmell J.C. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W.P., Zhou D., Ichim T.E., Strejan G.H., Xia X., Yang J., Huang X., Garcia B., White D., Dutartre P. Inhibitory feedback loop between tolerogenic dendritic cells and regulatory T cells in transplant tolerance. J. Immunol. 2003;170:1304–1312. doi: 10.4049/jimmunol.170.3.1304. [DOI] [PubMed] [Google Scholar]
- Nilsson J., Björkbacka H., Fredrikson G.N. Apolipoprotein B100 autoimmunity and atherosclerosis - disease mechanisms and therapeutic potential. Curr. Opin. Lipidol. 2012;23:422–428. doi: 10.1097/MOL.0b013e328356ec7c. [DOI] [PubMed] [Google Scholar]
- Painter M.W., Davis S., Hardy R.R., Mathis D., Benoist C., Immunological Genome Project Consortium Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J. Immunol. 2011;186:3047–3057. doi: 10.4049/jimmunol.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini L., Allan S.E., Battaglia M., Di Nunzio S., Alstad A.N., Levings M.K., Roncarolo M.G., Bacchetta R. STAT5-signaling cytokines regulate the expression of FOXP3 in CD4+CD25+ regulatory T cells and CD4+CD25- effector T cells. Int. Immunol. 2008;20:421–431. doi: 10.1093/intimm/dxn002. [DOI] [PubMed] [Google Scholar]
- Procaccini C., Matarese G. Regulatory T cells, mTOR kinase, and metabolic activity. Cell. Mol. Life Sci. 2012;69:3975–3987. doi: 10.1007/s00018-012-1058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccini C., De Rosa V., Galgani M., Abanni L., Calì G., Porcellini A., Carbone F., Fontana S., Horvath T.L., La Cava A., Matarese G. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Sauer S., Bruno L., Hertweck A., Finlay D., Leleu M., Spivakov M., Knight Z.A., Cobb B.S., Cantrell D., O’Connor E. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C., Hansmann L., Lassmann T., Balwierz P.J., Kawaji H., Itoh M., Kawai J., Nagao-Sato S., Suzuki H., Andreesen R., FANTOM consortium The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood. 2014;123:e68–e78. doi: 10.1182/blood-2013-02-486944. [DOI] [PubMed] [Google Scholar]
- Tandon P., Gallo C.A., Khatri S., Barger J.F., Yepiskoposyan H., Plas D.R. Requirement for ribosomal protein S6 kinase 1 to mediate glycolysis and apoptosis resistance induced by Pten deficiency. Proc. Natl. Acad. Sci. USA. 2011;108:2361–2365. doi: 10.1073/pnas.1013629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Adams J.Y., Penaranda C., Melli K., Piaggio E., Sgouroudis E., Piccirillo C.A., Salomon B.L., Bluestone J.A. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton A.M., Shevach E.M. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowinkel T., Mori M., Krieglstein C.F., Russell J., Saijo F., Bharwani S., Turnage R.H., Davidson W.S., Tso P., Granger D.N., Kalogeris T.J. Apolipoprotein A-IV inhibits experimental colitis. J. Clin. Invest. 2004;114:260–269. doi: 10.1172/JCI21233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukmanovic-Stejic M., Zhang Y., Cook J.E., Fletcher J.M., McQuaid A., Masters J.E., Rustin M.H., Taams L.S., Beverley P.C., Macallan D.C., Akbar A.N. Human CD4+ CD25hi Foxp3+ regulatory T cells are derived by rapid turnover of memory populations in vivo. J. Clin. Invest. 2006;116:2423–2433. doi: 10.1172/JCI28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukmanovic-Stejic M., Agius E., Booth N., Dunne P.J., Lacy K.E., Reed J.R., Sobande T.O., Kissane S., Salmon M., Rustin M.H., Akbar A.N. The kinetics of CD4+Foxp3+ T cell accumulation during a human cutaneous antigen-specific memory response in vivo. J. Clin. Invest. 2008;118:3639–3650. doi: 10.1172/JCI35834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A.J., Zabalawi M., Owen J.S., Shah D., Grayson J.M., Major A.S., Bhat S., Gibbs D.P., Jr., Thomas M.J., Sorci-Thomas M.G. Apolipoprotein A-I modulates regulatory T cells in autoimmune LDLr-/-, ApoA-I-/- mice. J. Biol. Chem. 2010;285:36158–36169. doi: 10.1074/jbc.M110.134130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., Neale G., Vogel P., Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Manzotti C.N., Liu M., Burke F., Mead K.I., Sansom D.M. CD86 and CD80 differentially modulate the suppressive function of human regulatory T cells. J. Immunol. 2004;172:2778–2784. doi: 10.4049/jimmunol.172.5.2778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.