Figure 4.
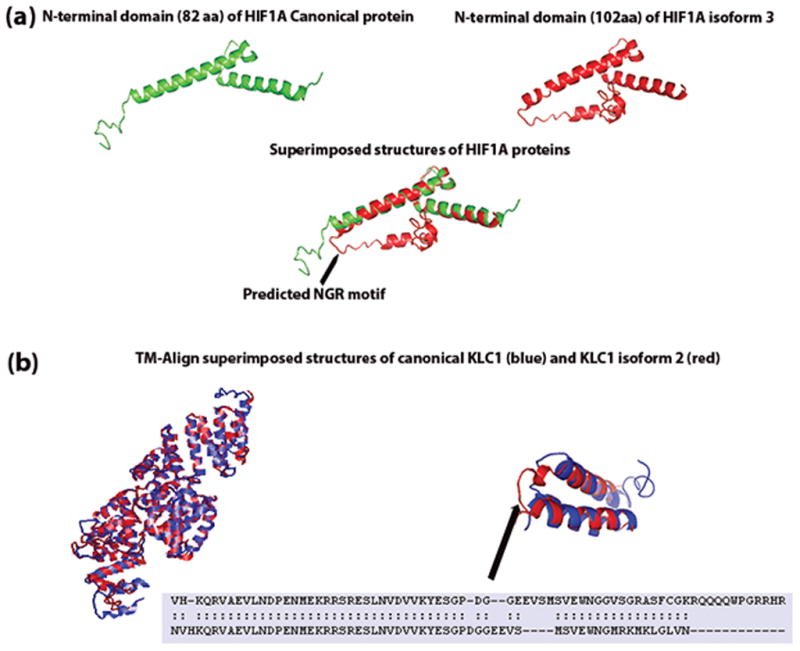
I-TASSER predicted structures of (a) first domain of HIF1A canonical and isoform 3 proteins. RMSD between the two predicted structures was 1.88 Å. The arrow points to the predicted NGR motif unique to HIF1A isform 3. (b) I-TASSER predicted structures of KLC1 canonical, and the isoform 2 proteins were aligned and superimposed using TM-align. Even though the two protein sequences differ at the c-terminal end, the superimposed structures reveal no structural differences. The overall RMSD between the two structures was 2.52 Å. Superimposed structures of the c-terminal ends (500–573 aa for KLC1 canonical protein and 500–560 aa for KLC1 isoform c) showed structural alignment differences at PDGG residues (534–537 aa).
