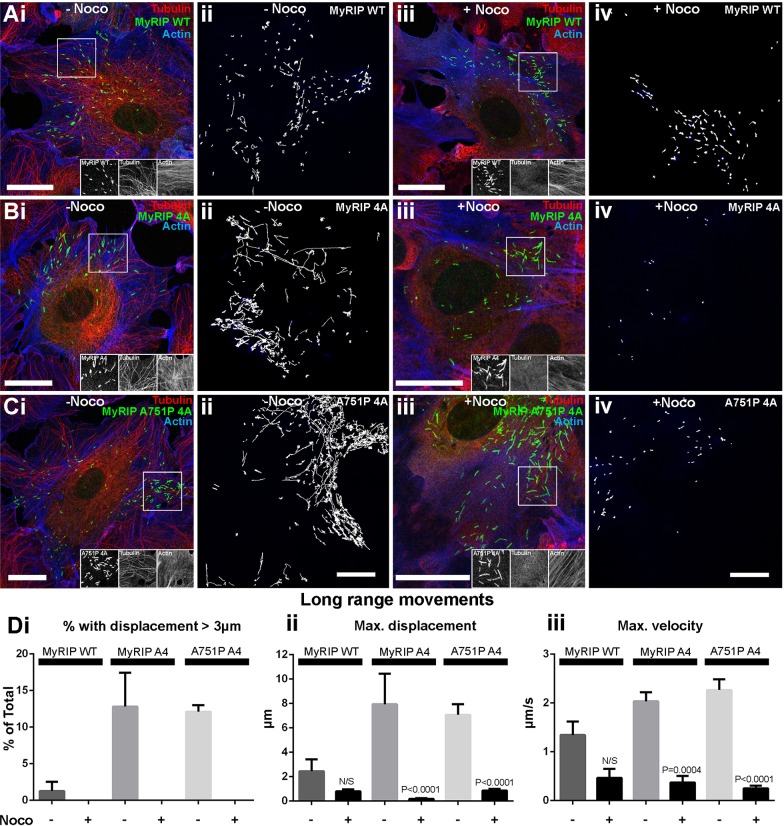
Fig. 6.
Microtubule disruption abolishes long-range movements of WPBs carrying actin-binding-defective EGFP–MyRIP mutants. (Ai–Ci) Representative confocal immunofluorescence images of individual HUVECs expressing EGFP fusion proteins of MyRIP WT (Ai), MyRIP 4A (Bi) or MyRIP A751P 4A (Ci) exposed to vehicle (0.1% DMSO, 20 min: −Noco) and labelled with specific antibodies to EGFP (green), α-tubulin (red) and actin (phalloidin; blue). Scale bars: 20 µm. (Aii–Cii) Representative examples of the x-y trajectories of individual WPBs in control (−Noco) HUVECs expressing EGFP fusion proteins of MyRIP WT (Aii), MyRIP 4A (Bii) or MyRIP A751P 4A (Cii). Number of cells imaged and trajectories detected were: MyRIP WT, n=7 cells, 1007 trajectories; MyRIP 4A, n=3 cells, 448 trajectories; MyRIP A751P 4A, n=8 cells, 1667 trajectories. Scale bar: 10 µm for Aii-Cii (shown in Cii). (Aiii–Ciii) Representative confocal immunofluorescence images of individual HUVECs expressing the same EGFP fusion proteins and stained as described in Ai–Ci, but after treatment with nocodozole (1 µM, 20 min: +Noco). Scale bars: 20 µm. (Aiv–Civ) Representative examples of the x-y trajectories of individual WPBs in +Noco treated HUVECs expressing EGFP fusion proteins of MyRIP WT (Aiv), MyRIP 4A (Biv) or MyRIP A751P 4A (Civ). Number of cells imaged and trajectories detected were: MyRIP WT, n=3 cells, 497 trajectories; MyRIP 4A, n=7 cells, 125 trajectories; MyRIP A751P 4A, n=10 cells, 392 trajectories. Scale bar: 10 µm for Aiv-Civ (shown in Civ). (Di–iii) Mean±s.e.m. values for the fraction of WPBs with movements greater than 3 µm in length (Di), maximum displacement from start of recording (Dii) and maximum velocity (Diii) determined from the detected trajectories of the WPBs in control (−Noco) or nocodozole (+Noco) treated cells expressing the MyRIP fusion proteins as indicated. Number of cells imaged and trajectories detected in −Noco and +Noco treated cells are as described above. P values were calculated with a one-way ANOVA using Tukey multiple comparisons test.