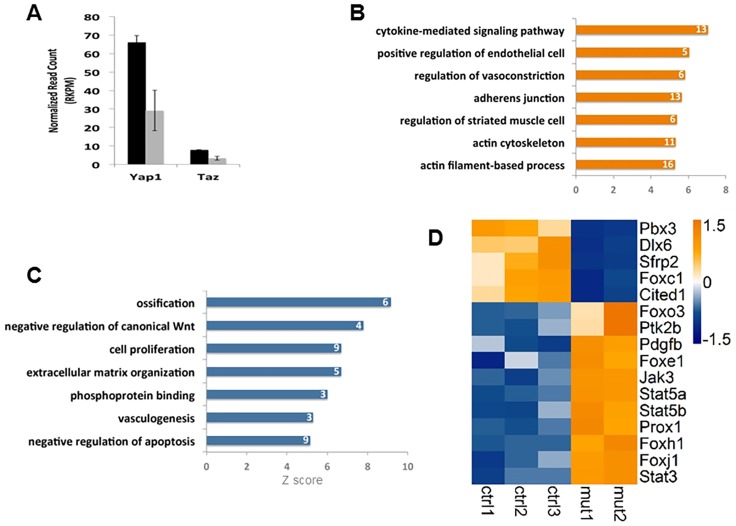
Fig. 5.
The regulation of multiple signals by the Hippo pathway in CNC-derived cells. RNA-Seq analysis was performed by using mandibular tissues from E10.5 control embryos and Wnt1Cre Taz; Yap dCKO mutants. (A) RNA-Seq analysis indicates that Yap and Taz expression levels (indicated by reads) decreased in Wnt1Cre; Taz; Yap dCKO embryos compared with control embryos. (B) Gene ontology analysis shows genes that are upregulated in Wnt1Cre; Taz; Yap dCKO embryos compared with control embryos, which includes genes that regulate adherens junctions, vasoconstriction and the cytoskeleton. (C) Genes that are downregulated in Wnt1Cre; Taz; Yap dCKO embryos compared with control embryos, which includes genes that regulate cell proliferation and vasculogenesis and that negatively regulate canonical Wnt. Numbers in bars in B and C indicate gene number of each gene ontology term. (D) Heat map of RNA-Seq data shows that, compared with controls, Wnt1Cre; Taz; Yap dCKO mutants downregulate expression of Foxc1 and upregulate expression of Foxe1, Prox1, Pdgfb and Jak-Stat genes, including Jak3, Ptk2b, Stat3, Stat5a and Stat5b.