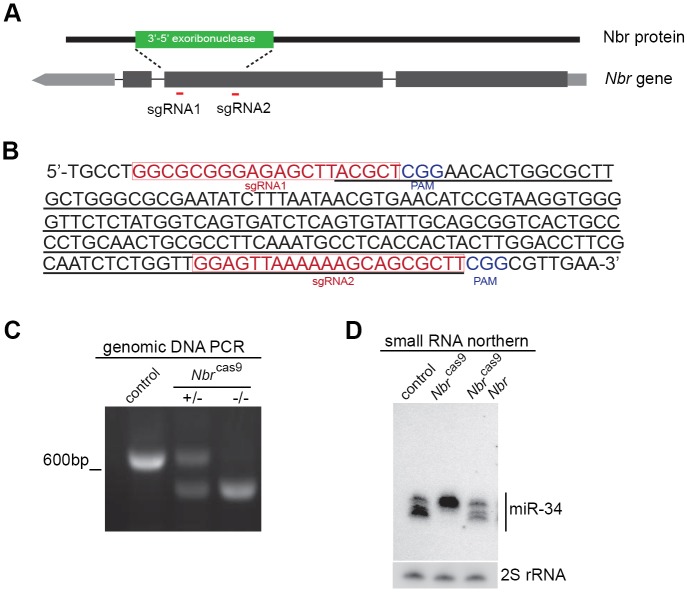
Fig. 2.
Generation of a new Nbr loss-of-function allele. (A) The Nbr locus and guiding RNAs used to make Nbrcas9. To produce an Nbr loss-of-function allele using the CRISPR/Cas9 method, sgRNA1 and sgRNA2 (red lines) were used to induce site-specific deletion of the 3′-to-5′ exoribonuclease domain of Nbr. (B) Part of the Nbr genomic sequence illustrating features of the CRISPR/Cas9-mediated Nbrcas9 loss-of-function allele. sgRNAs and protospacer adjacent motifs (PAMs) are highlighted. Nucleotides deleted are underlined. Resulting Nbr mutant flies were backcrossed to the control homogeneous background for five generations to ensure background clearance. (C) PCR analysis confirms that Nbrcas9 is a deletion allele. DNAs were from whole flies. Genotypes: control (5905), Nbrcas9/+ and Nbrcas9/cas9. (D) Small RNA northern blot confirms that Nbrcas9 is a loss-of-function allele. Whereas control flies expressed miR-34 normally, showing three major mature forms, Nbrcas9 flies lacked the smaller isoforms with an accumulation of the top isoform, reflecting a trimming defect. Expression of a wild-type Nbr transgene in Nbrcas9 flies restored all miR-34 isoforms, indicating a functional rescue. RNAs were from whole flies. Genotypes: control (5905), Nbrcas9/cas9 (Nbrcas9) and pUAST-Nbr, Nbrcas9/cas9; GeneSwitch-tubulin-GAL4 (Nbrcas9 Nbr).