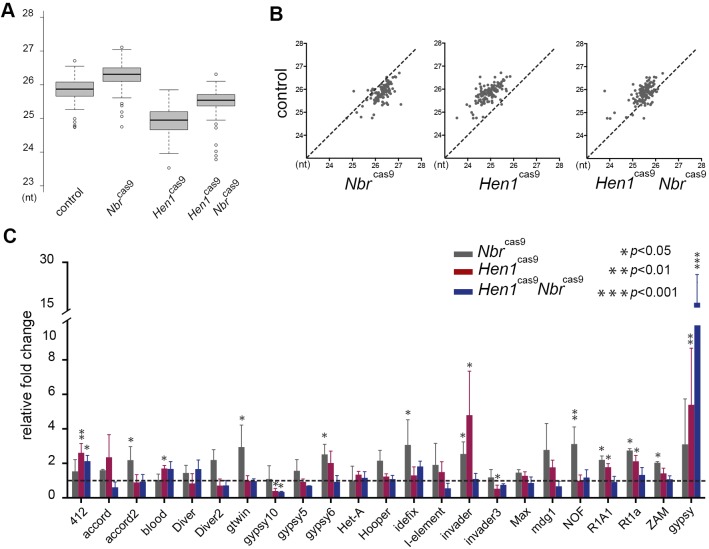
Fig. 5.
Antagonistic roles of Nbr and Hen1 in piRNA pathways. (A,B) Box plots (A) and scatter plots (B) showing that Nbr and Hen1 modulate piRNA length. Whereas lack of Nbr lengthened piRNAs, lack of Hen1 shortened them. Flies lacking both Nbr and Hen1, however, showed restoration of the piRNA profile towards that of controls. Nbrcas9 versus control (5905), P<2.2×10−16; Hen1cas9 versus control, P<2.2×10−16; Hen1cas9 Nbrcas9 versus control, P<2.2×10−16; Wilcoxon signed-rank test. piRNA sequencing and analysis of RNA from ovaries of 3-day-old animals of the indicated genotypes. Genotypes: Nbrcas9/cas9 (Nbrcas9), Hen1cas9/cas9 (Hen1cas9) and double mutant. (C) Nbr and Hen1 are functionally relevant in repressing TEs. qRT-PCR analysis was used to determine the expression status of TEs among different genotypes. Whereas lack of either Nbr or Hen1 caused upregulation of select transposons, flies mutant for both Nbr and Hen1 exhibited a clear reversion in that the majority of the desilenced TEs became repressed, the only exceptions being 412 and gypsy. Mean±s.d., n=3 independent experiments; Student's t-test. RNAs were from ovaries. Genotypes as in A.