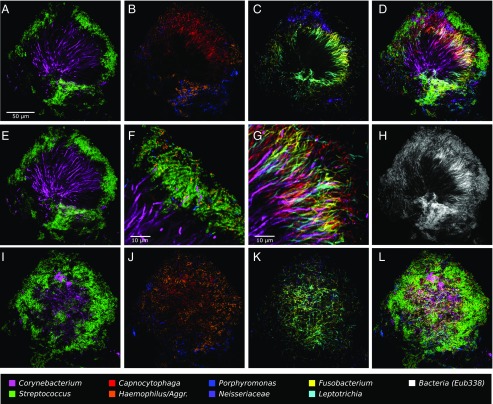
Fig. 3.
A hedgehog structure in plaque showing spatial organization of the plaque microbiome. Plaque was hybridized with a set of 10 probes each labeled with a different fluorophore. Each panel shows the superposition of several of these individual fluorophore channels. A–D and F–H show a single focal plane near the center of the structure, with two to three fluorophore channels shown in each of A–C and all nine specific probes superimposed in D. (E) Maximum intensity projection of three planes, representing a total of ∼2 μm of thickness, to visualize the continuity of Corynebacterium filaments from the center toward the edge of the structure. F is a detailed view of corncob structures. G is a detailed view of mixed filaments. H shows the fluorophore channel corresponding to the universal bacterial probe, showing that the specific probes (D) identify most of the cells that hybridize to the universal probe. I–L show a second focal plane near the periphery of the structure. Fluorophore channels shown correspond to the following genera in the figure: (A, E, and I) Corynebacterium and Streptococcus; (B and J) Capnocytophaga, Porphyromonas, and Haemophilus/Aggregatibacter; (C and K) Fusobacterium, Leptotrichia, and Neisseriaceae; (D and L) all nine specific probes; (F) Corynebacterium, Streptococcus, Porphyromonas, and Haemophilus/Aggregatibacter; (G) Corynebacterium, Fusobacterium, Leptotrichia, and Capnocytophaga; and (H) Bacteria. The plaque sample was fixed in 2% (wt/vol) paraformaldehyde, stored in 50% (vol/vol) ethanol, and spread onto the slide in 50% (vol/vol) ethanol in preparation for FISH.