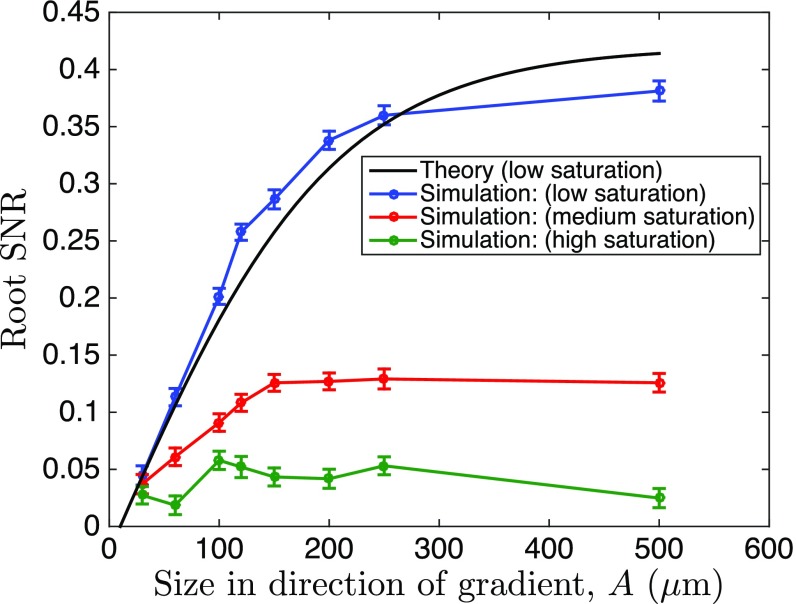
Fig. 4.
Saturation of signaling responses reduces the maximum SNR in the simulation of multicellular gradient sensing. To investigate effects of signaling saturation, we simulated a multicellular stochastic LEGI model, using a spatially resolved implementation of the stochastic simulation algorithm (Supporting Information), and measured the simulated organoid bias at different saturation levels. Shown is the square root of the SNR (mean response squared over its variance), where for the response we take the deviation of the species activated by the local and suppressed by the global messenger from its mean value. This is the closest equivalent to and to the bias measure in Fig. 3 and Fig. S6. For the theory and the simulations, we set molecules per cell volume per cell diameter, cells, and molecules per cell volume. We contrasted the theory and the simulations by matching the low saturation curves for small organoid sizes. The effects of saturation in activation/suppression of the response by the messengers result in earlier saturation of the SNR curves, as expected.