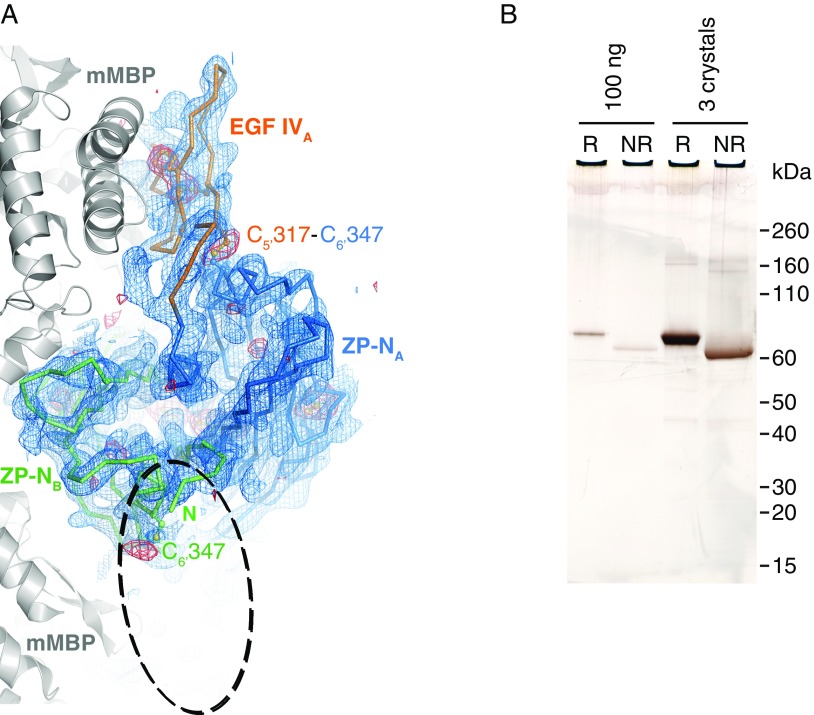
Fig. S2.
Analysis of crystallized mMBP-UMODpXR. (A) Top view of the ZP-N dimer, shown as a Cα-trace together with EGF IV, and mMBP molecules involved in crystal packing (gray cartoon). Blue mesh is a simulated-annealing composite omit map contoured at 1.0 σ. Red mesh is an anomalous difference map calculated with density-modified phases from the initial mMBP MR solution and contoured at 3.0 σ. The difference map, which is not biased by the structure of the UMOD moiety, shows peaks for all visible Cys residues. There is no density for the EGF IV domain in molecule B, which should be located in the area of the dashed ellipse. EGF IV in molecule A is stacked between ZP-N and mMBP, whereas in molecule B, it is protruding into the solvent, which allows it to be more flexible. Accordingly, the C-terminal part of mMBP in molecule B also shows a high degree of flexibility. (B) SDS/PAGE comparison of purified and crystallized material shows that mMBP-UMODpXR in the crystals is not degraded. Furthermore, the relative shift in migration between samples run in reducing (R) and nonreducing (NR) conditions, as well as the absence of additional bands, indicates that there is not even partial reduction of disulfide bonds. Crystallization at pH 7.0 and/or data collection in the presence of electron-scavenging ascorbate or oxidized glutathione did not result in clearly resolvable electron density for EGF IV in chain B. This observation excludes that flexibility is due to loss of the C5′–C6′ interdomain disulfide bond. Because molecule B represents 50% of the material in the crystal, protein degradation or disulfide bond reduction should be readily detectable by SDS/PAGE.