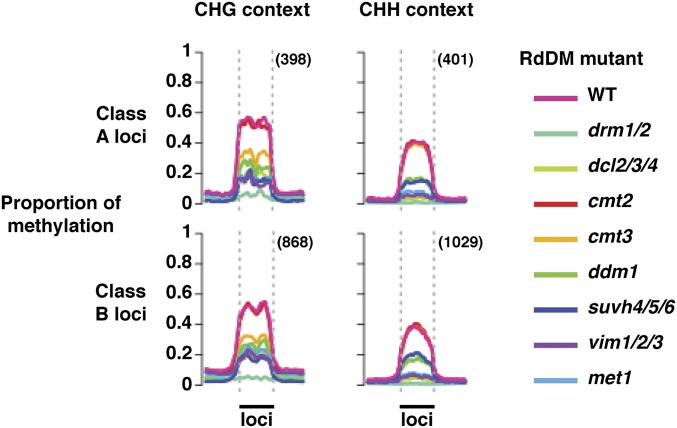
Fig. 2.
DNA methylation of mobile sRNA-associated loci (class A, direct; B, indirect) is reduced most strongly by drm1/2 mutation. The plots show the mean proportion of DNA methylation across class A and B loci in mutants of RNA-directed DNA methylation pathway components and wild-type (WT) plants. The legend (Right) indicates which color represents each mutant. Data are shown separately for non-CG (CHG and CHH) sequence contexts, which had the strongest association with mobile sRNAs. The proportion of methylation was calculated per base (between 0 and 1, unmethylated to fully methylated) for all loci within a class using published MethylC-seq data from RdDM mutants (31). Loci were then normalized to the same size, and the mean proportion of DNA methylation was calculated across them. The profiles of the mean proportion of methylation across the size-normalized loci are plotted between the dashed vertical lines (indicated by the solid black bars labeled “loci”). Mean proportions of methylation in flanking DNA, 4 kb upstream and downstream of the loci, are indicated to the left and right of the dashed vertical lines, respectively. Total numbers of loci assessed are given in parentheses.