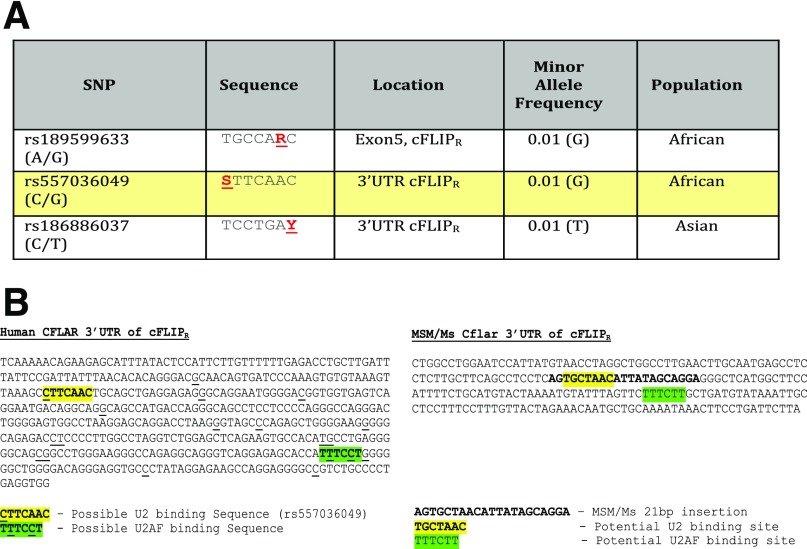
Fig. S2.
SNPs identified from the 1000 Genomes Project that may alter putative U2 snRNP binding regions near the splice site that distinguishes cFLIPL from cFLIPR in humans. (A) Table showing data mined from the 1000 Genomes database from exon 5 and the 3′ UTR following exon 5 from the CFLAR gene. The sequence surrounding the SNP is provided where R = A/G, S = C/G, and Y = C/T. The rs557036049 SNP is highlighted in yellow because the underrepresented allele (G) is not preferred by the U2 snRNP and thus may decrease the binding efficacy of this protein, which is crucial for initiating splicing—leading to the production of cFLIPL. We postulate that fewer splicing events may result in greater levels of cFLIPR transcript per protein. (B) Human sequence data from the Ensembl genome browser or sequencing data from MSM mouse genomic DNA showing a portion of the 3′ UTR of exon 5. The highlighted yellow sequence identifies a possible U2 biding site, and the highlighted green sequence identifies a possible U2AF binding sequence proximal to the U2 binding site. In humans, there are polymorphisms in both putative U2 and U2AF sites that may regulate binding of these proteins (and possibly also of the spliceosome). MSM mice introduce a possible U2 binding sequence proximal to a putative U2AF binding site. This insertion is absent in B6 mice. We postulate that this insertion may allow for the recruitment of the spliceosome complex to either enhance splicing or impede the transcription of cFLIPR, either way leading to decreased production of cFLIPR and relatively elevated expression of cFLIPL.