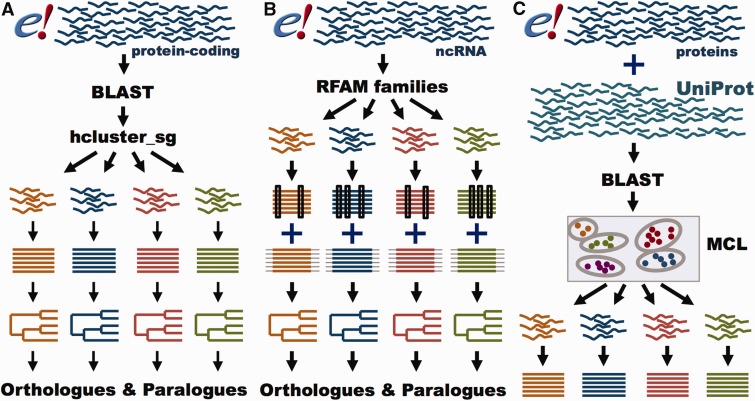
Figure 4.
GeneTree and Ensembl Protein Family pipelines. (A) GeneTree pipeline for protein-coding genes. For each protein-coding gene in Ensembl, a representative protein is used. BLAST scores are provided to hcluster_sg for grouping the sequences into gene families. The proteins are aligned with MCoffee or MAFFT and a phylogenetic tree is built with TreeBeST. Finally, orthologues and paralogues are inferred from the tree. (B) GeneTree pipeline for ncRNA genes. Short ncRNA genes in Ensembl are grouped according to their RFAM classification. Both Infernal and PRANK alignments are used to build several phylogenetic trees that are merged into a final model with TreeBeST. Finally, orthologues and paralogues are inferred from the tree. (C) Ensembl Protein Family pipeline. All proteins in Ensembl and all metazoan proteins in UniProt are used. BLAST scores are fed into MCL to group the sequences by their similarity. The proteins are aligned with MAFFT.