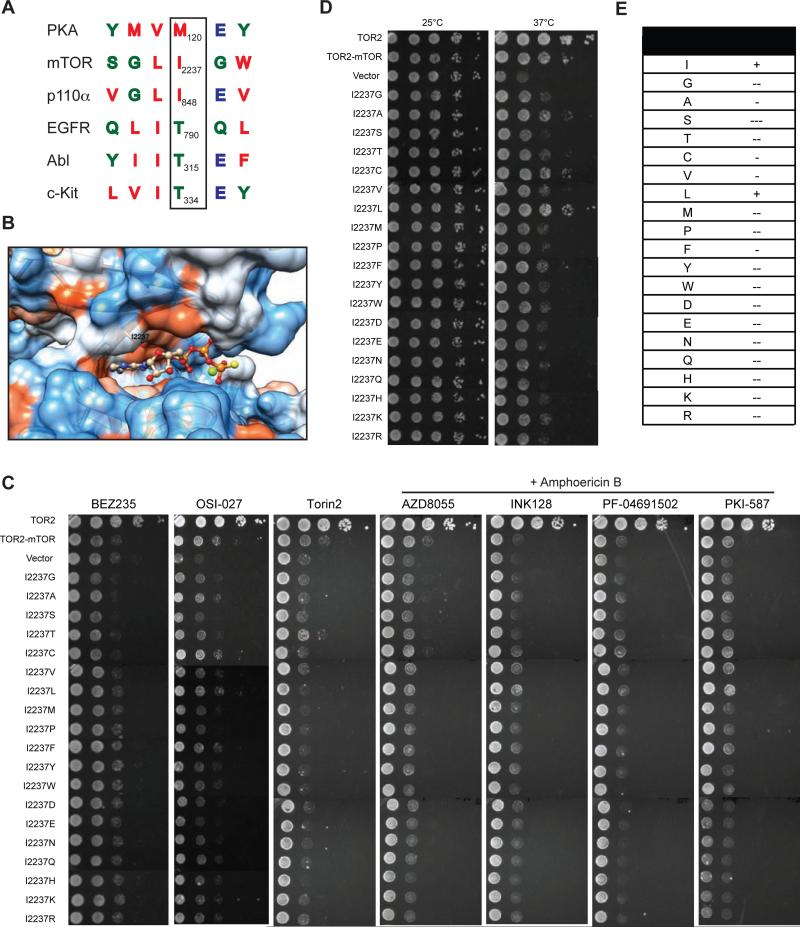
Figure 3. Mutational analysis of the gatekeeper residue in mTOR kinase.
(A) Sequence alignment of the gatekeeper site for PKA, c-Kit, EGFR, ABL, p110-PI3Kα, and mTOR. Arrowhead marks the gatekeeper residue.
(B) Electrostatic model of the ATP-binding pocket of mTOR kinase (PDB ID code 4JSP). The I2237 position is as indicated. Atom is colored as follows: N, blue; O, red; P, orange; S, yellow; Mg+2, green. Surface representation is as follows: hydrophobic residue, red; neutral residue, white; hydrophilic residue, blue.
(C) tor2-dg cells expressing WT or mutant TOR2-mTOR were serially diluted by 10-fold and assayed for drug sensitivity on SC-leucine plates containing BEZ235, OSI-027, or Torin2, or AZD8055, BEZ235, INK128, PF-04691502 or PKI-587 in the presence of amphotericin B.
(D) tor2-dg cells expressing WT or mutant TOR2-mTOR were serially diluted by 10-fold and assayed for cell growth at different temperatures. Vector and TOR2 plasmids were used as a negative and positive control, respectively.
(E) Summary of gatekeeper mutations and their effects on mTOR kinase function. “+”: normal function; “−”: minor defect; “−−”: moderate defect, and “−−−”: severe defect.