Abstract
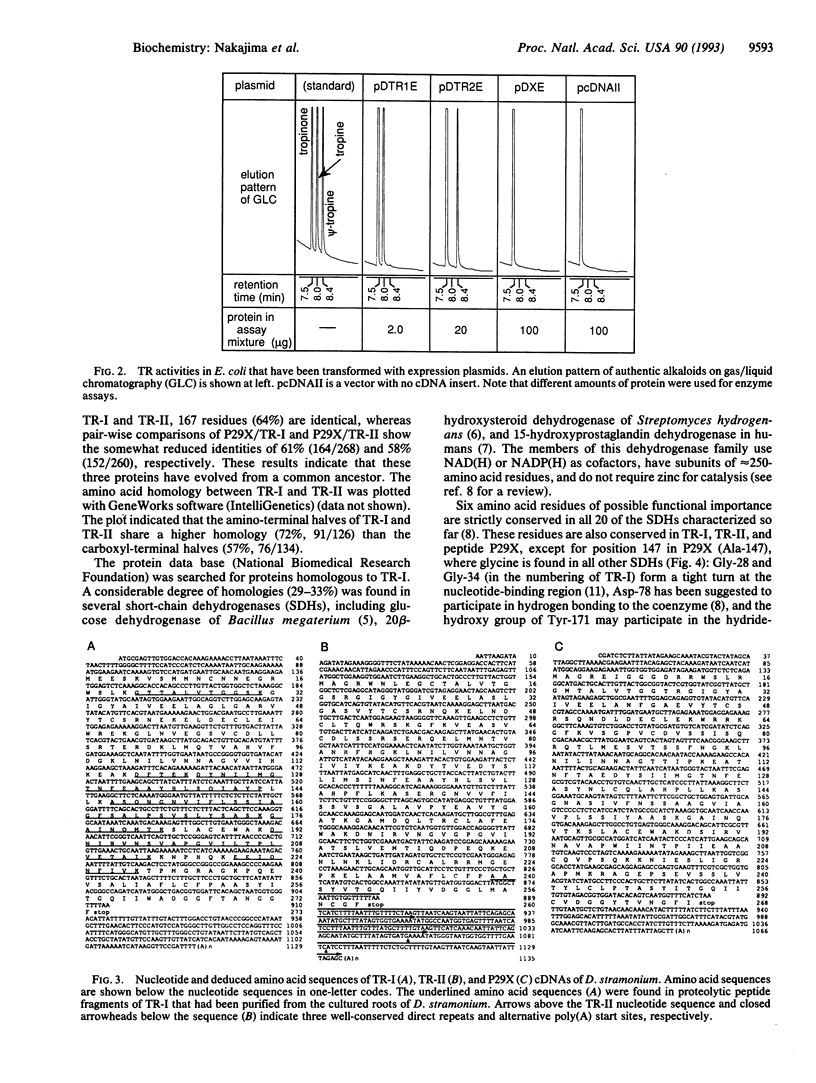
In the biosynthetic pathway of tropane alkaloids, tropinone reductase (EC 1.1.1.236) (TR)-I and TR-II, respectively, reduce a common substrate, tropinone, stereospecifically to the stereoisomeric alkamines tropine and pseudotropine (psi-tropine). cDNA clones coding for TR-I and TR-II, as well as a structurally related cDNA clone with an unknown function, were isolated from the solanaceous plant Datura stramonium. The cDNA clones for TR-I and TR-II encode polypeptides containing 273 and 260 amino acids, respectively, and when these clones were expressed in Escherichia coli, the recombinant TRs showed the same strict stereospecificity as that observed for the native TRs that had been isolated from plants. The deduced amino acid sequences of the two clones showed an overall identity of 64% in 260-amino acid residues and also shared significant similarities with enzymes in the short-chain, nonmetal dehydrogenase family. Genomic DNA-blot analysis detected the TR-encoding genes in three tropane alkaloid-producing solanaceous species but did not detect them in tobacco. We discuss how the two TRs may have evolved to catalyze the opposite stereospecific reductions.
Full text
PDF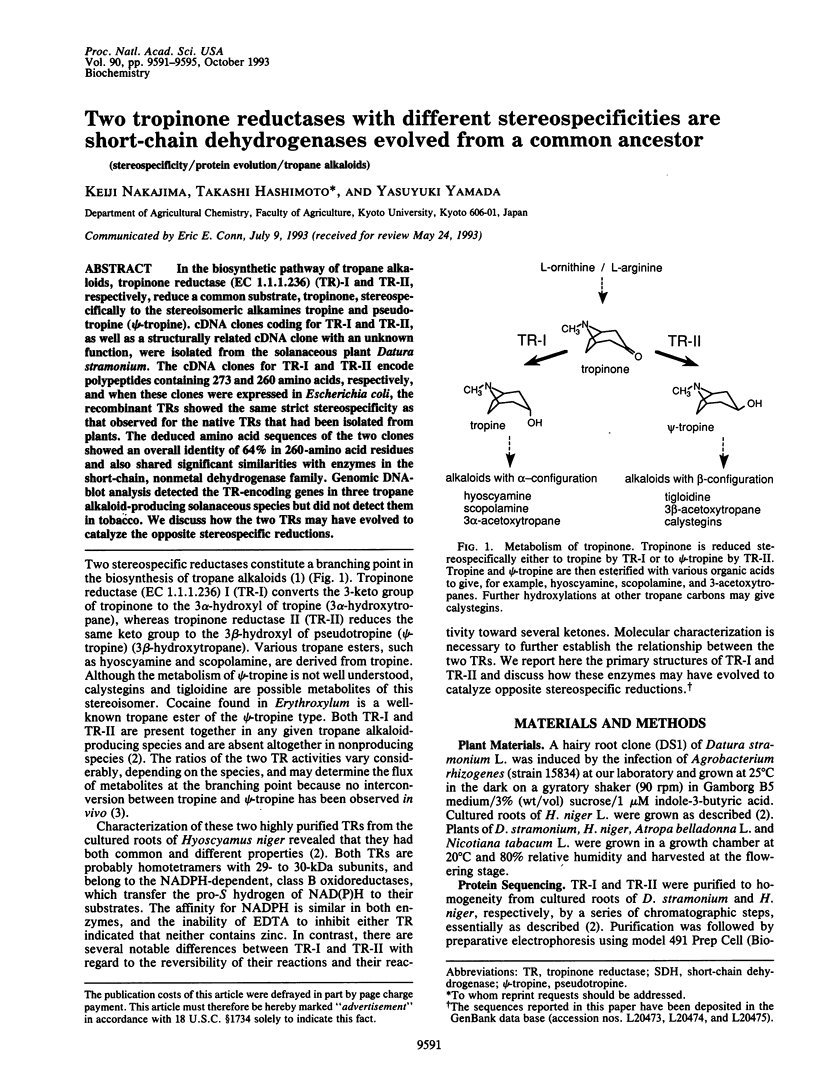



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albalat R., González-Duarte, Atrian S. Protein engineering of Drosophila alcohol dehydrogenase. The hydroxyl group of Tyr152 is involved in the active site of the enzyme. FEBS Lett. 1992 Aug 24;308(3):235–239. doi: 10.1016/0014-5793(92)81282-q. [DOI] [PubMed] [Google Scholar]
- Clarke A. R., Atkinson T., Holbrook J. J. From analysis to synthesis: new ligand binding sites on the lactate dehydrogenase framework. Part II. Trends Biochem Sci. 1989 Apr;14(4):145–148. doi: 10.1016/0968-0004(89)90147-3. [DOI] [PubMed] [Google Scholar]
- Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., Orr J. C. Three-dimensional structure of holo 3 alpha,20 beta-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Nakajima K., Ongena G., Yamada Y. Two Tropinone Reductases with Distinct Stereospecificities from Cultured Roots of Hyoscyamus niger. Plant Physiol. 1992 Oct;100(2):836–845. doi: 10.1104/pp.100.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie S., Doi S., Yorifuji T., Takagi M., Yano K. Nucleotide sequencing and characterization of the genes encoding benzene oxidation enzymes of Pseudomonas putida. J Bacteriol. 1987 Nov;169(11):5174–5179. doi: 10.1128/jb.169.11.5174-5179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jany K. D., Ulmer W., Fröschle M., Pfleiderer G. Complete amino acid sequence of glucose dehydrogenase from Bacillus megaterium. FEBS Lett. 1984 Jan 2;165(1):6–10. doi: 10.1016/0014-5793(84)80003-4. [DOI] [PubMed] [Google Scholar]
- Krook M., Marekov L., Jörnvall H. Purification and structural characterization of placental NAD(+)-linked 15-hydroxyprostaglandin dehydrogenase. The primary structure reveals the enzyme to belong to the short-chain alcohol dehydrogenase family. Biochemistry. 1990 Jan 23;29(3):738–743. doi: 10.1021/bi00455a021. [DOI] [PubMed] [Google Scholar]
- Leete E. Recent developments in the biosynthesis of the tropane alkaloids. Planta Med. 1990 Aug;56(4):339–352. doi: 10.1055/s-2006-960979. [DOI] [PubMed] [Google Scholar]
- Marekov L., Krook M., Jörnvall H. Prokaryotic 20 beta-hydroxysteroid dehydrogenase is an enzyme of the 'short-chain, non-metalloenzyme' alcohol dehydrogenase type. FEBS Lett. 1990 Jun 18;266(1-2):51–54. doi: 10.1016/0014-5793(90)81504-h. [DOI] [PubMed] [Google Scholar]
- Persson B., Krook M., Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991 Sep 1;200(2):537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- Sloane D. L., Leung R., Craik C. S., Sigal E. A primary determinant for lipoxygenase positional specificity. Nature. 1991 Nov 14;354(6349):149–152. doi: 10.1038/354149a0. [DOI] [PubMed] [Google Scholar]
- Taguchi H., Ohta T. D-lactate dehydrogenase is a member of the D-isomer-specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the D-lactate dehydrogenase gene of Lactobacillus plantarum. J Biol Chem. 1991 Jul 5;266(19):12588–12594. [PubMed] [Google Scholar]
- Thatcher D. R. The complete amino acid sequence of three alcohol dehydrogenase alleloenzymes (AdhN-11, AdhS and AdhUF) from the fruitfly Drosophila melanogaster. Biochem J. 1980 Jun 1;187(3):875–883. doi: 10.1042/bj1870875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin L., DeLuca V., Ibrahim R. K., Brisson N. Molecular characterization of two plant flavonol sulfotransferases. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1286–1290. doi: 10.1073/pnas.89.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]




