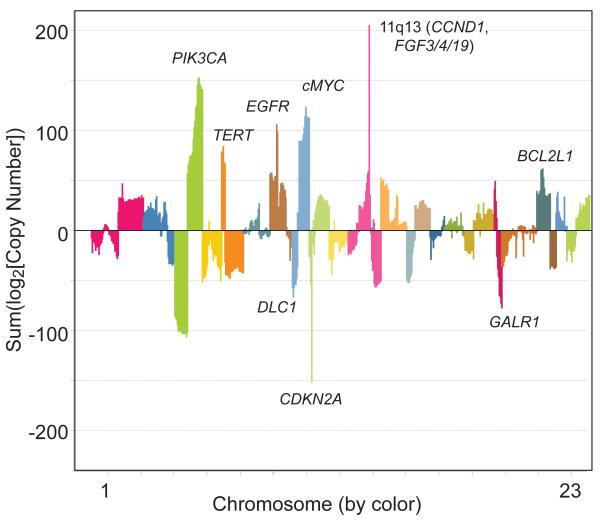
Figure 1. Meta-analysis of copy number alterations in publicly available aCGH and NGS HNSCC datasets.
Copy number data from aCGH or NGS data sets from Stransky [17], Peng [79], Agrawal [14] and the HNSCC TCGA [15] data (n = 500) sets were pooled for a meta-analysis of copy number alterations in HNSCC. For each sample, the log2 value of the copy number alteration was calculated and samples were summed on a gene by gene level [e.g. to calculate the overall copy number alteration across these data sets, the following summation was performed: log2(Copy number EGFR_sample1) + log2(Copy number EGFR_sample2)…etc. + log2(Copy number EGFR_sample500)]. Thus, the most recurrently altered genes with high level amplifications or deletions from all genetic subsets of HNSCC are highlighted in the plot. Representative and established HNSCC oncogenes and tumor suppressors are listed next to respective chromosome alterations.