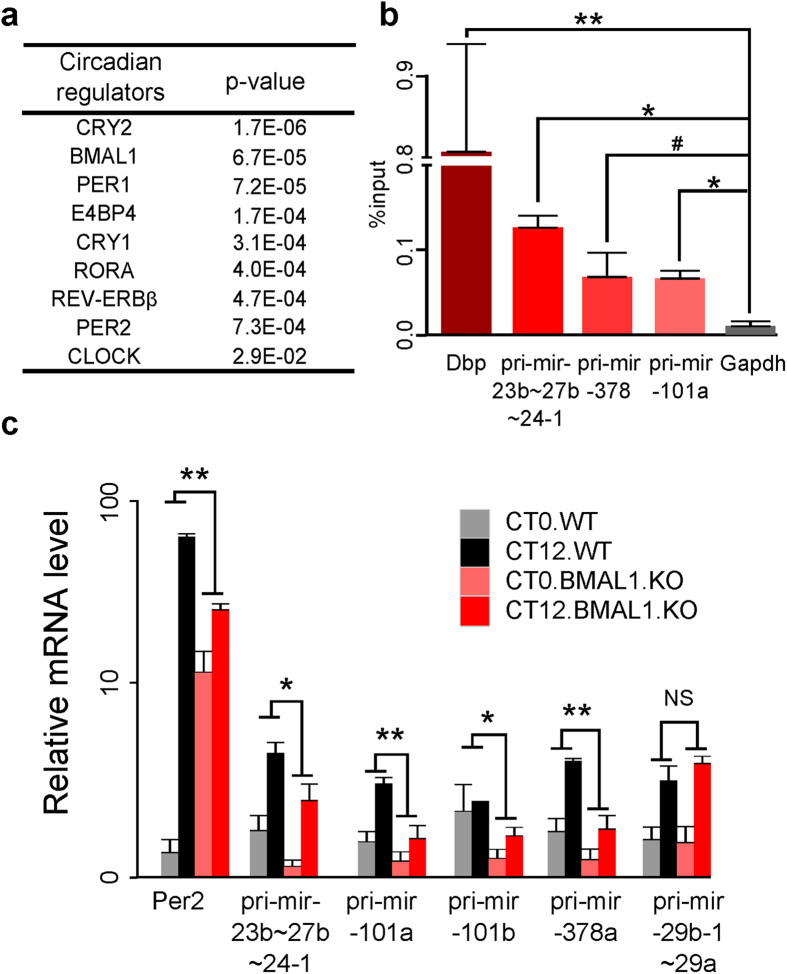
Figure 2. The regulation of circadian miRNA primary transcripts by circadian regulators.
(a) circadian regulator binging sites are enriched on the promoters of the circadian oscillating miRNA primary transcripts comparing to all miRNA primary transcripts. Enrichment p-values were calculated by proportion test. (b) BMAL1 ChIP-PCR showed that the promoter regions of pri-mir-23b~27b~24-1, pri-mir-378, and pri-mir-101a are bound by BMAL1/CLOCK. Gapdh is used as the negative control, while Dbp is used as the positive control. **represents Student’s t-test (unpaired, two-sided) p-value < 0.01, *represents Student’s t-test (unpaired, two-sided) p-value < 0.05. #represents Student’s t-test (unpaired, two-sided) p-value < 0.1 (marginally significant). (c) qPCR analysis of circadian miRNA primary transcripts of liver-specific Bmal1 cKO mice and wild-type mice. Per2 is used as the positive control. The y-axis has an exponential scale. Two-way ANOVA using circadian time (CT0 and CT12) and genotype (Bmal1 cKO vs. wild-type) as two factors were applied to assess the statistical significance. **represents ANOVA p-value for genotype < 0.01, *represents ANOVA p-value for genotype < 0.05, NS stands for not significant p-value.