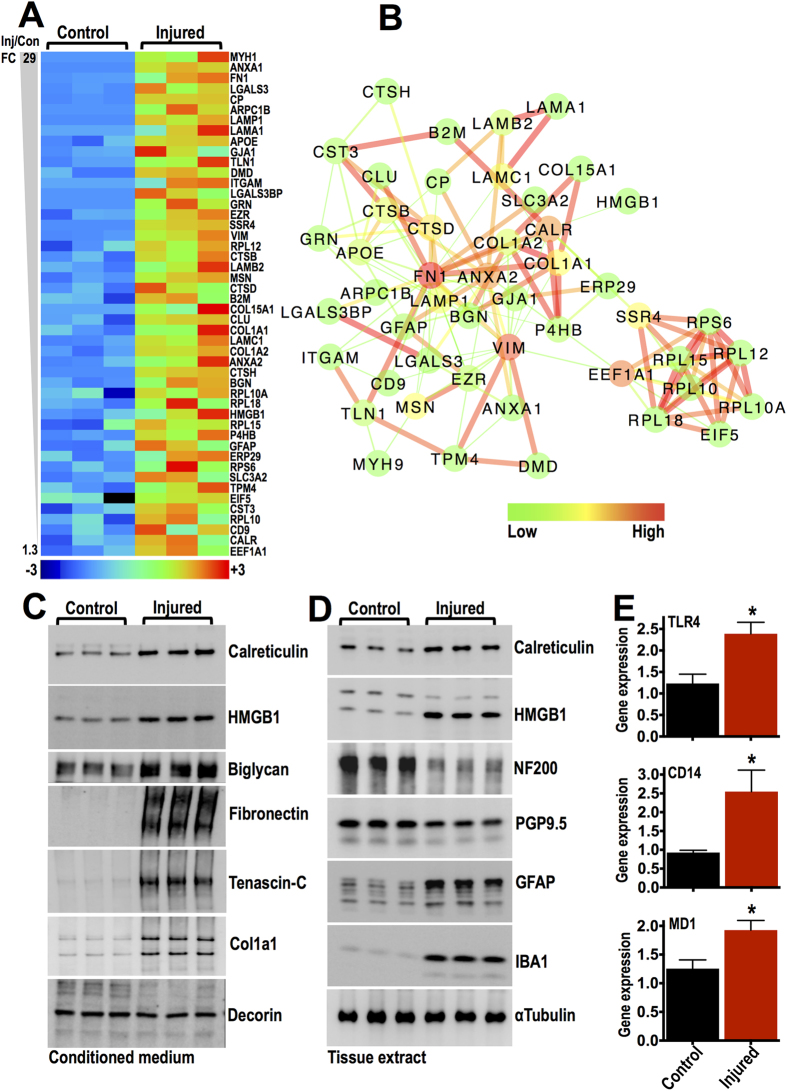
Figure 4. Identification of persistently upregulated bioactive molecules.
(A) Heat-map displays 48 upregulated proteins found in proteomics analysis of guanidine extracts 8 weeks post-injury. Proteins are listed on the heat-map according to their fold change (FC, Injured/Controls). These molecules were also upregulated at the transcript level 5 weeks post-injury as detected by microarray analysis (see reference 29). (B) Network analysis (StringDB, v9.1) of the 48 genes upregulated both at the mRNA and protein level. Node color indicates betweeness centrality while edge color indicates interaction score based on the predicted functional links between nodes (green: low values; red: high values). (C,D) Immunoblotting of conditioned medium (C) and tissue extracts (D) derived from either uninjured control or injured T10 spinal cord explants cultured for 24 hours in plain DMEM culture medium. The endogenous TLR4 ligands HMGB1, biglycan, fibronectin (EDA fragments) and tenascin-C are present in the conditioned medium. Calreticulin and HMGB1 were also upregulated in tissue extracts. (E) Relative mRNA expression of TLR4, CD14 and MD1 measured by TaqMan quantitative PCR in either control or injured T10 spinal cord explants, 7 days post-injury. ACTB served as the housekeeping gene. N = 3 animals per group; * P ≤ 0.05, two-tailed t-test.