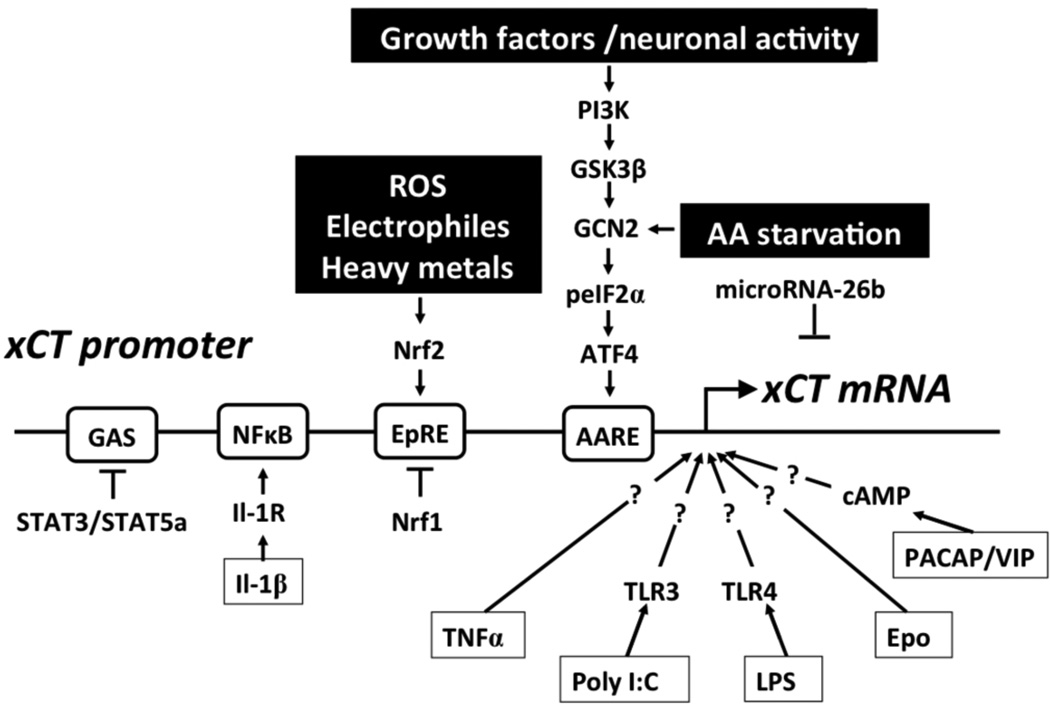
Figure 2. Transcriptional regulation of xCT expression.
A variety of stimuli, e.g. electrophiles, heavy metals, and reactive oxygen species (ROS), lead to activation of the nuclear factor NF-E2-related factor 2 (Nrf2), which binds to the electrophile response element (EpRE) within the xCT promoter region and activates transcription. Nrf1 inhibits the action of Nrf2. Amino acid (AA) starvation leads to phosphorylation of eIF2a (peIF2α), which via GCN2 leads to the translational up-regulation of the transcription factor activating transcription factor 4 (ATF4). ATF4 activates the transcription of xCT by binding to the amino acid response element (AARE) contained in the xCT promoter. Alternatively, growth factors and neuronal activity can induce xCT promoter activity via phosphoinositol-3 kinase (PI3K), glycogen synthase-3β (GSK-3β) and the GCN2/eIF2α/ATF4 module. STAT3/STAT5A negatively regulate the xCT promoter activity via a GAS element. Interleukin-1β (IL-1β) activates the xCT promoter by binding to the interleukin 1 receptor (Il-1R) and NF-κB. Tumor necrosis factor α (TNFα) and erythropoietin (EPO) increase the transcription of xCT through unknown signaling pathways. Bacterial lipopolysaccharides (LPS) and polyinosinic-polycytidylic acid (Poly I:C) activate the xCT promoter presumably via TLR4 and TLR3, respectively. PACAP and VIP increase cAMP and thereby activate xCT transcription. MicroRNA-26b directly targets xCT mRNA.