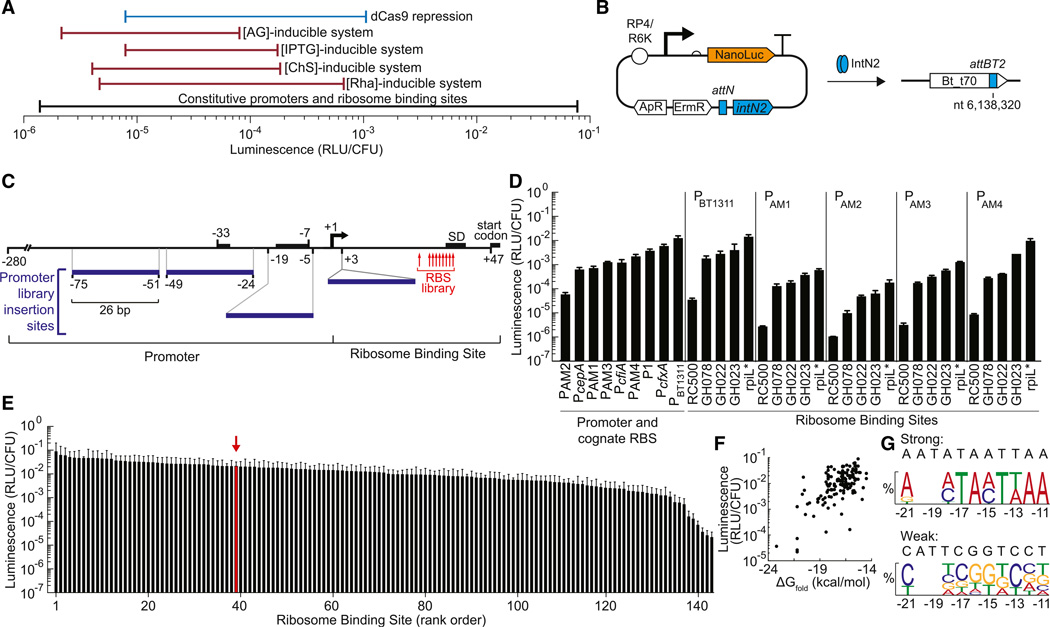
Figure 1. Genetic Parts to Control Expression in B. thetaiotaomicron.
(A) The ranges of gene expression are shown for the different gene regulation systems developed in this manuscript. AG, arabinogalactan; CS, chondroitin sulfate; IPTG, isopropyl beta-D-1-thiogalactopyranoside; Rha, rhamnose; dCas9, catalytically inactive Cas9 endonuclease
(B) Tyrosine integrase IntN2 catalyzes stable integration of pNBU2-based expression constructs into one of two attBT2 sites in the B. thetaiotaomicron genome (Wang et al., 2000). The two attBT2 sites (attBT2-1 at nt 6,217,227 and attBT2-2 at nt 6,138,320) are in the 3′ ends of tRNASer genes (BT_t71 and BT_t70, respectively). ApR, ampicillin resistance cassette; ErmR, erythromycin resistance cassette; RP4, origin of transfer; R6K, origin of replication; NanoLuc, luciferase
(C) Constitutive promoters and ribosome binding sites for the construction of gene expression libraries. The putative −33 and −7 regions of the PBT1311 promoter, the Shine-Delgarno sequence, and the start codon are indicated by black boxes. Numbers below the black boxes represent nucleotide locations relative the PBT1311 transcription start site. The 26 bp sequence introduced in the PAM promoters is shown as blue boxes (see also Figure S1). Numbers at the edges of the blue boxes indicate the PBT1311 nucleotides replaced or the insertion site within the promoter. The location of residues randomized in the rpiL* RBS library are indicated with red arrows (for library A: nt −14, −13, −12; for library B: nt −21, −18, −15; and for library C: nt −17, −16, −11; nt numbering is relative to the translation start site).
(D) Activity was measured for a set of constitutive promoters and their cognate RBSs. For PAM1, PAM2, PAM3, PAM4, the BT1311 RBS was used. Furthermore, PBT1311, PAM1, PAM2, PAM3, and PAM4 were combined with RBSs of varying strengths. Gene expression was measured using a luciferase reporter (NanoLuc) and reported as relative light units/colony forming unit (RLU/CFU).
(E) Three large RBS libraries were constructed and combined with promoter PBT1311. For reference, the parent rpiL* RBS is indicated with a red arrow. The sequences of the RBSs are provided in Table S1. For (D) and (E), error bars represent the SD of three independent biological replicates made on separate days.
(F) The strength of each RBS was compared to the predicted free energy of folding for the mRNA (ΔGfold).
(G) A consensus strong RBS and weak RBS were generated for the rpiL* RBS library using frequency logos that included the 11 strongest and 11 weakest RBSs (residue locations are stated relative to the translation start site). Frequency logos were constructed by comparing the frequency of each nucleotide at each position in that group with the frequency of that nucleotide in that position in the full library. Position −20 and −19 were not randomized and are not shown in the frequency logos.