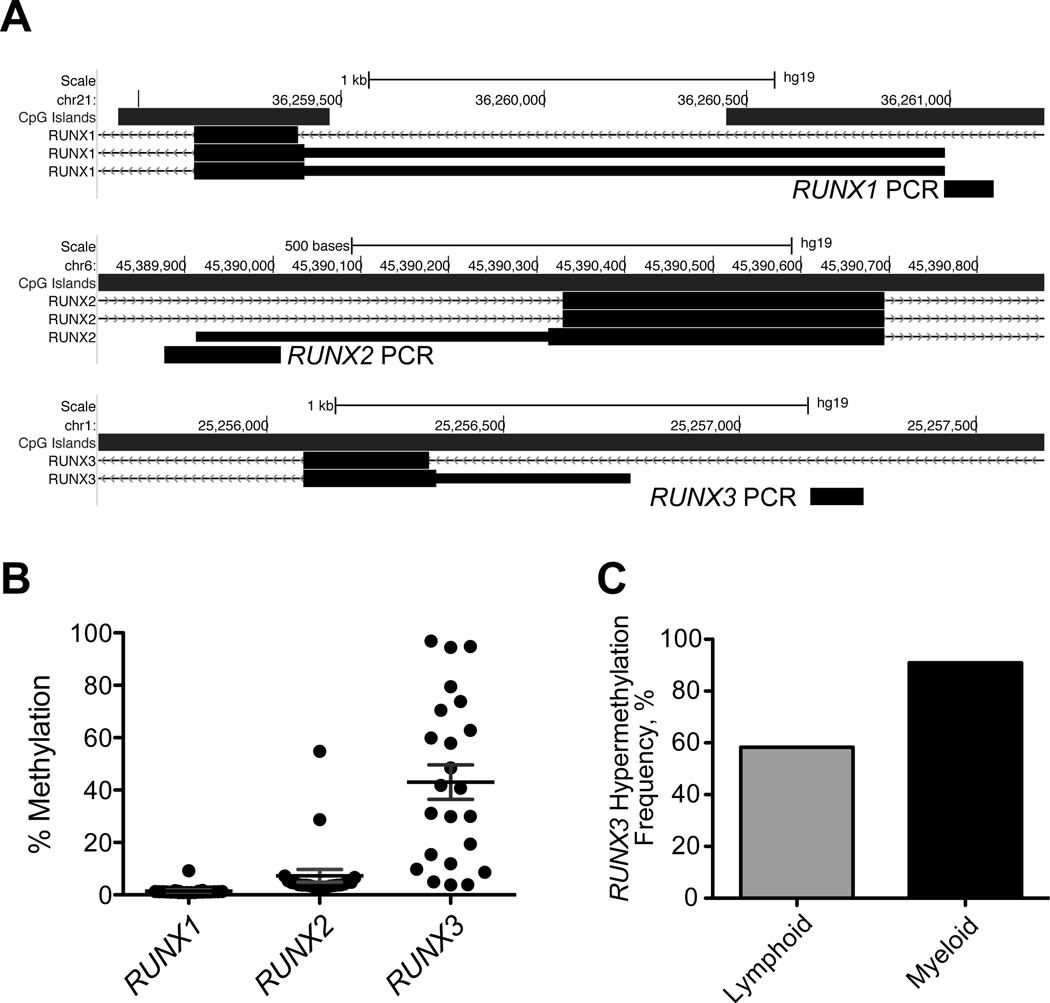
Figure 1.
Gene promoter DNA methylation analysis of RUNX1, RUNX2, and RUNX3 in leukemia cell lines. (A) Schematic representation of bisulfite PCR primers for downstream pyrosequencing analysis. The generated PCR amplicons were within −500bp to +200bp from the transcription start site (TSS) of the alternative promoter (P2) of each gene, and the methylation density of 2 to 3 CpG sites per amplicon was quantified and averaged. (B) DNA methylation density of RUNX1, RUNX2, and RUNX3 in 23 leukemia cell lines. Note that only RUNX3 shows frequent promoter DNA hypermethylation (defined as 15% measured DNA methylation and above). (C) Differential frequency of RUNX3 promoter DNA hypermethylation in lymphoid versus myeloid leukemias.