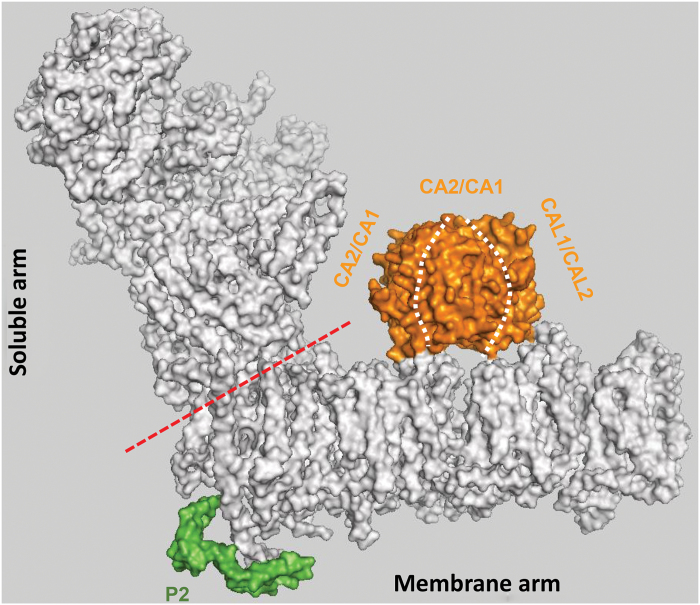
Fig. 1.
A hypothetical 3D-structural model of Arabidopsis CI. The proton-pumping CI enzyme is a large bi-partite membranous protein assembly, which has a central role in the production of cellular energy in bacteria and mitochondria. CI has a characteristic L-shaped structure with the hydrophobic arm embedded in the membrane and the hydrophilic peripheral arm facing into the cytoplasm (bacteria) or the mitochondrial matrix. The bacterial enzyme is relatively simple, containing 14 different subunits, while the mitochondrial enzymes contain extra subunits. Sequence data for the different plant CI subunits is according to Meyer (2012). Homology models were constructed individually and fitted according to the electron density map of the bovine CI enzyme (Shimada et al., 2014). The approximate positions of the CA-related subunits were fitted based on the low-resolution negative stained map of Arabidopsis CI (Sunderhaus et al., 2006). The solvent accessible surface for the Arabidopsis CI enzyme was visualized in Pymol (v1.3). Different subunits that are shared between the plant CI and its related bacterial and mitochondrial enzymes are shown in grey; extra subcomplex I domains of the membrane arm, including the matrix-facing CA module (orange) and the P2-domain (green), are highlighted. The suggested CA module is an assembly with triplets containing either CA1-CA1-CAL1 (or CAL2) or CA2-CA2-CAL1 (or CAL2) subunits.