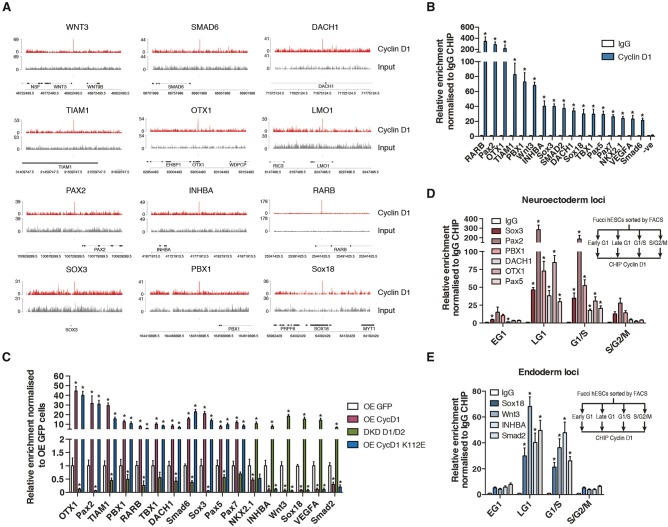
Figure 4.
Cyclin D1 binds to its developmental target genes in a cell cycle-dependent manner. (A) Representative Cyclin D1-binding regions close to developmental genes spanning 50 kb upstream of and 50 kb downstream from each ChIP-seq peak. Cyclin D1 binding was identified by ChIP-seq analysis of endogenous Cyclin D1 in hESCs. (B) Cyclin D1 ChIP-qPCR at genomic regions close to selected developmental loci that were identified as Cyclin D1-binding sites by ChIP-seq. Significant differences calculated by t-test are marked. (C) Cyclin D1 overexpression induces its neuroectodermal target genes while reducing its endoderm target genes. Gene expression changes of Cyclin D target genes in Cyclin D1-overexpressing cells, CycD1-K112E-overexpressing cells, and Cyclin D1/Cyclin D2 double-knockdown cells expressing shRNAs compared with control cells expressing GFP. Significant differences compared with GFP overexpression and calculated by t-test are marked. (D,E) Cell cycle-dependent binding of Cyclin D1 to neuroectoderm genes (D) and endoderm genes (E). Cyclin D1 ChIP was performed on Tra-1–60-positive FACS-sorted Fucci hESCs, and then qPCR was performed to detect the genes denoted. Significant differences compared with G2/M phase and calculated by t-test are marked. All data are shown as mean ± SD. n = 3.