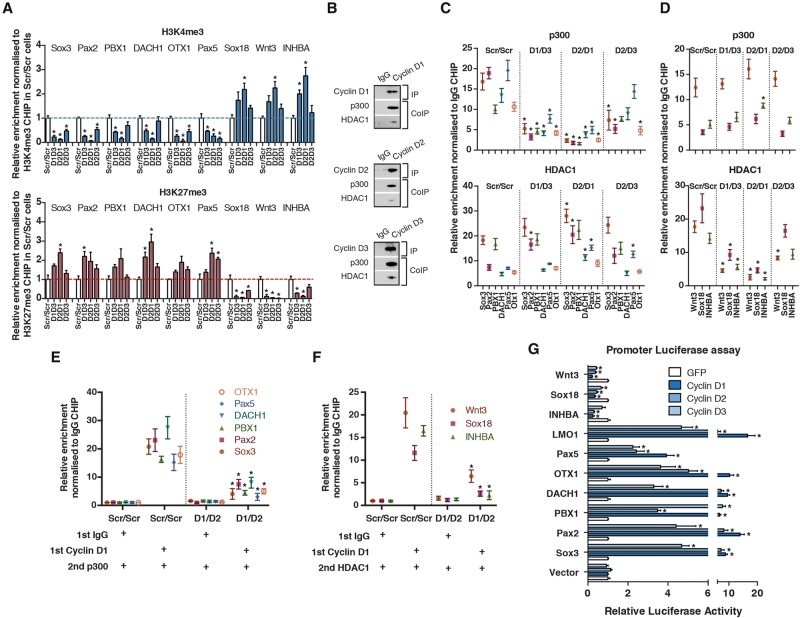
Figure 5.
Cyclin Ds interact with transcriptional coregulators to control germ layer specification genes. (A) Cyclin D expression promotes the deposition of active histone mark H3K4me3 on neuroectoderm genes and repressive histone mark H3K27me3 on endoderm genes. ChIP of histone H3K4me3 and H3K27me3 was performed on Tra-1–60-positive Cyclin D double-knockdown hESCs, and then qPCR was performed to detect genomic regions corresponding to selected key developmental genes. hESCs expressing Scramble shRNA (Scr/Scr) were used as a control. The dashed line indicates no change with respect to Scr/Scr ChIP. Significant differences compared with Scr/Scr and calculated by t-test are marked. (B) Cyclin D1–D3 interact with p300 and HDAC1 on chromatin of hESCs. Cyclin Ds were immunoprecipitated from chromatin fractions and analyzed for the presence of p300 and HDAC1 by Western blot. (C,D) Cyclin D proteins recruit transcriptional coregulators to loci controlling germ layer specification. ChIP of coactivator p300 and corepressor HDAC1 was performed on Tra-1–60-positive Cyclin D double-knockdown hESCs, and then qPCR was performed to detect genomic regions corresponding to developmental genes regulating neuroectoderm (C) and endoderm (D) specification. hESCs expressing Scramble shRNA (Scr/Scr) were used as a control. Significant differences compared with Scr/Scr and calculated by t-test are marked. (E,F) Cyclin D is necessary to recruit p300 to neuroectoderm loci and HDAC1 to endoderm loci in hESCs. Sequential ChIP was carried out in Scr/Scr and Cyclin D1/D3 double-knockdown cells by first performing a Cyclin D1 ChIP followed by p300 or HDAC1 ChIP on neuroectoderm (E) and endoderm (F) loci. Significant differences compared with Scr/Scr and calculated by t-test are marked. (G) Cyclin Ds can induce transcriptional activity of neuroectoderm genes and repress transcriptional activity of endoderm genes. A luciferase expression construct containing promoter regions of the developmental genes with Cyclin D1-binding sites was transfected together with Cyclin D1–3 and analyzed for luciferase activity 48 h after transfection. Renilla luciferase was used as an internal control during transfections. Significant differences compared with GFP overexpression and calculated by t-test are marked. All data are shown as mean ± SD. n = 3.