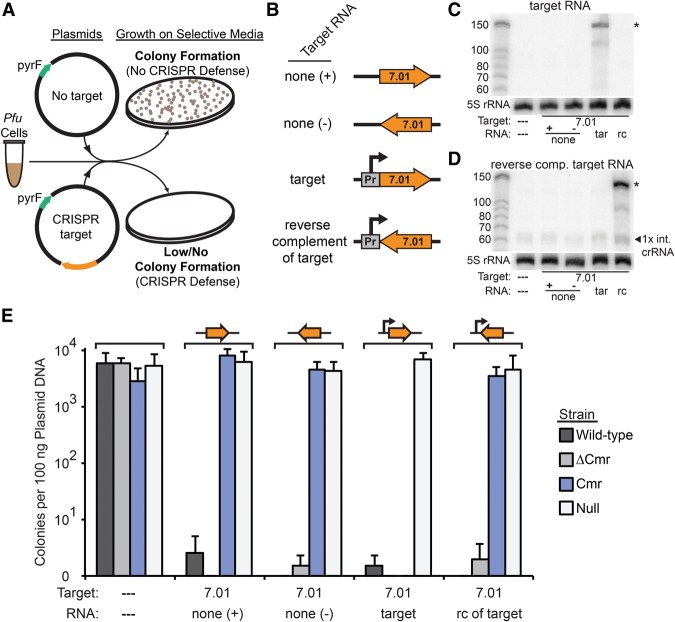
Figure 1.
The P. furiosus (Pfu) Cmr system silences plasmid DNA in a transcription-dependent manner. (A) Plasmid challenge assay. In the absence of CRISPR–Cas defense, plasmid infection results in colony formation (growth of cells transformed with a plasmid containing the pyrF gene in the absence of uracil). (B) Target sequence transcription configuration of the various plasmids. The orientation of the 7.01 crRNA target sequence relative to the promoter and plasmid backbone is shown. Plasmids were designed for no transcription (none +/−), transcription of a target RNA complementary to the endogenous 7.01 crRNA (target [tar]), or transcription of an RNA that is not complementary to the 7.01 crRNA (reverse complement of target [rc]). (C,D) Northern analysis of expression of the 7.01 target RNA (C) or the reverse complement RNA (D). The left lane is Decade marker RNA (Life Technologies). All other lanes contain 10 µg of total RNA isolated from P. furiosus TPF20 strains bearing plasmids configured to express the indicated RNA. The primary product is indicated with an asterisk. Blots were also probed for 5S rRNA as a loading control. (E) Colonies produced by infection with five plasmids in wild-type (three endogenous CRISPR–Cas systems; dark gray), ΔCmr (lacking the Cmr system; medium gray), Cmr (Cmr only; blue), and null (no system; light gray) strains. The presence and orientation of crRNA 7.01 target sequence on the plasmids are indicated by an orange arrow above the graph and a dashed line or 7.01 below the graph. The presence of the promoter for target region transcription on the plasmid is indicated by a line arrow above the graph, and the presence of a target region transcript is indicated below the graph as a dashed line, “none,” “target,” and “rc of target.” Colony numbers are plotted with the standard deviation in nine replicates indicated by error bars.