Figure 2.
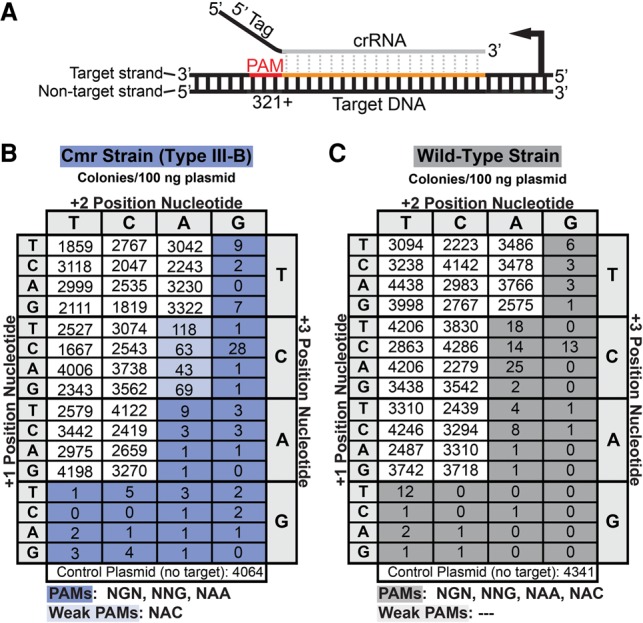
The Cmr system in P. furiosus (Pfu) uses a PAM to distinguish invader from host. (A) The location of tested PAM sequences (red) relative to the target sequence in the DNA (orange) and aligned crRNA. Dotted gray lines indicate complementarity between the crRNA guide region (gray) and DNA. (B,C) Results of plasmid infection assays for Cmr (blue) and wild-type (gray) strains, respectively. The plasmids contain the transcribed 7.01 target sequence and the indicated flanking (PAM) sequence. Colony numbers are the average of at least three replicates. Target-adjacent sequences that activated CRISPR–Cas targeting resulting in >100-fold reduction in colony numbers relative to negative control plasmid are shaded dark blue or gray. Sequences that conferred 30-fold to 100-fold reduction in colony numbers are shaded light blue or gray. The plasmid sequence downstream from the PAM (positions +4 to +8) in all plasmids is 5′-TTCCG-3′.
