Figure 3.
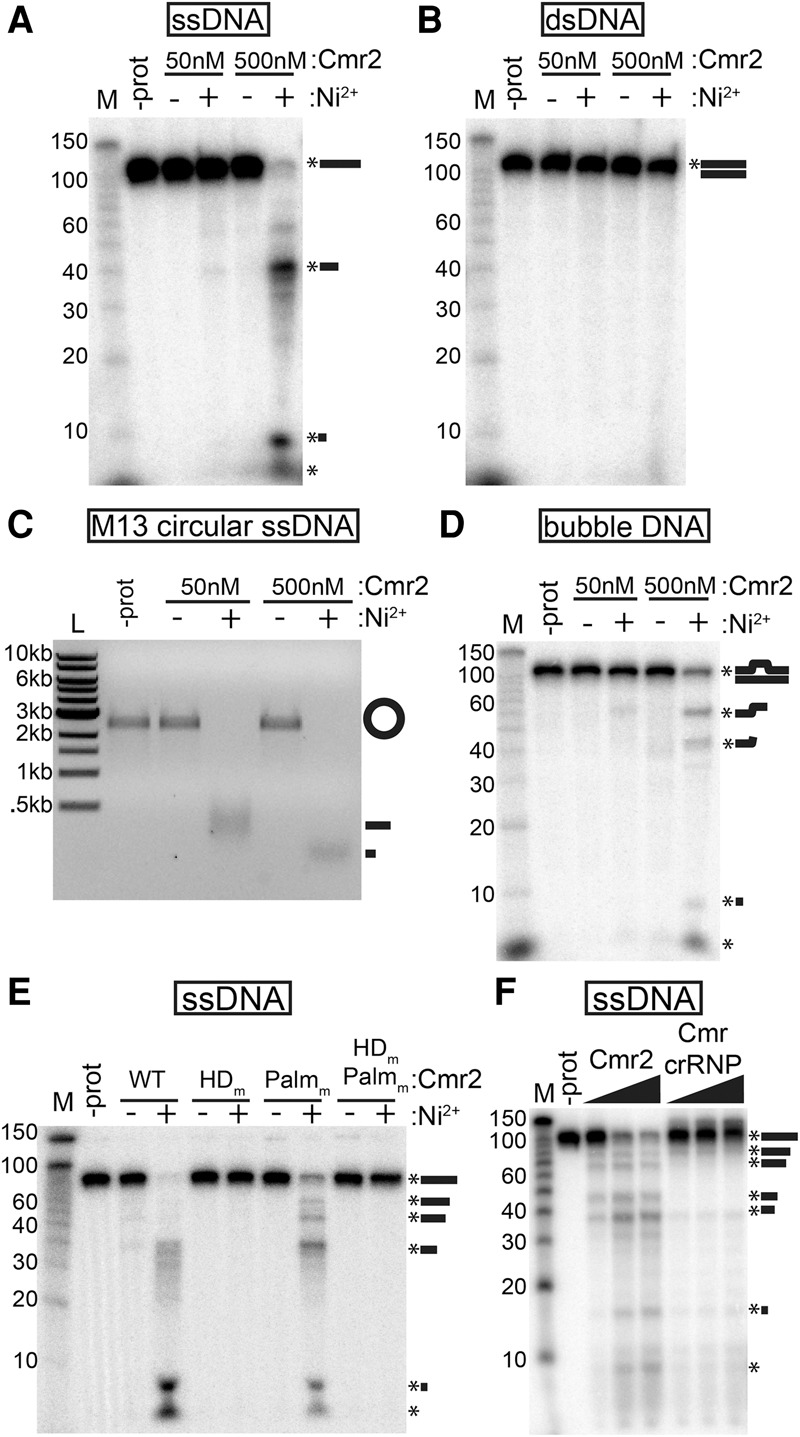
Cmr2 is a ssDNA endonuclease. (A–D) Substrate analysis. Recombinant Cmr2 (50 or 500 nM) was incubated with (+) or without (−) added NiCl2 (200 µM) in the presence of a 5′ radiolabeled ssDNA oligo (A), a 5′ radiolabeled dsDNA annealed from oligos (B), unlabeled circular ssM13 phage DNA (C), or a 5′ radiolabeled “bubble” DNA substrate (with a central single-stranded region) annealed from two partially complementary ssDNA oligos (D). (E) Mutant analysis. Wild-type (WT), HD domain putative active site mutant (HDm), Palm domain GGDD motif mutant (Palmm), or double-mutant (HDm,Palmm) Cmr2 proteins were incubated with 5′ radiolabeled ssDNA with (+) or without (−) added NiCl2. (F) Activity in the context of the Cmr complex. Increasing amounts (50–200 nM) of either recombinant Cmr2 or preformed Cmr crRNP complexes (Cmr1–6 + crRNA) were incubated with 5′ radiolabeled ssDNA oligo. Products were analyzed by denaturing PAGE (A,B,D–F) or agarose gel electrophoresis (C). Radiolabeled Decade markers (M) or New England Biolabs 1-kb ladder (L) were used for size estimation.
