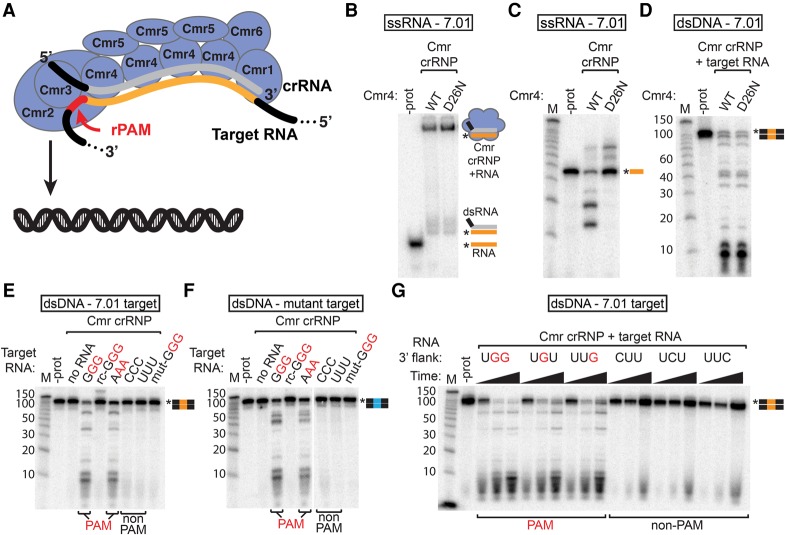
Figure 6.
Target RNAs containing a PAM sequence trigger Cmr dsDNA cleavage activity. (A) Model showing the Cmr complex (Cmr1–6 proteins and crRNA with the guide region in gray) activated by a target RNA containing a sequence recognized by crRNA (orange) and an rPAM (PAM sequence in the target RNA) sequence (red). The Cmr2 protein of the activated complex cleaves dsDNA. (B,C) Target RNA binding and cleavage by Cmr4 mutant complexes. 5′ radiolabeled 7.01 target RNA was incubated with Cmr crRNPs containing wild-type (WT) or mutant (D26N) Cmr4, and products were analyzed by native PAGE (B) or denaturing PAGE (C). (D) DNA cleavage activity of the Cmr4 mutant complex. Wild-type and Cmr4 mutant Cmr complexes were incubated with target RNA and 5′ radiolabeled dsDNA containing the crRNA 7.01 target sequence, and products were analyzed by denaturing PAGE. (E–G) Analysis of RNA PAM (rPAM) sequences. Cmr crRNPs were incubated with target RNAs containing the indicated flanking sequences (as well as the crRNA 7.01 target sequence) and with 5′ radiolabeled dsDNA substrates containing the crRNA 7.01 target sequence (E,G, orange) or a mutated target sequence (F, blue). Products were analyzed by denaturing PAGE. For G, reactions were analyzed at 5, 15, and 60 min. Flanking sequences corresponding to PAMs (identified in vivo) are indicated in red. Graphical representations of substrates are shown with the 7.01 target (orange) and mutant 7.01 target (blue) sequences and the radiolabeled strands (asterisks) indicated.