Fig. 3.
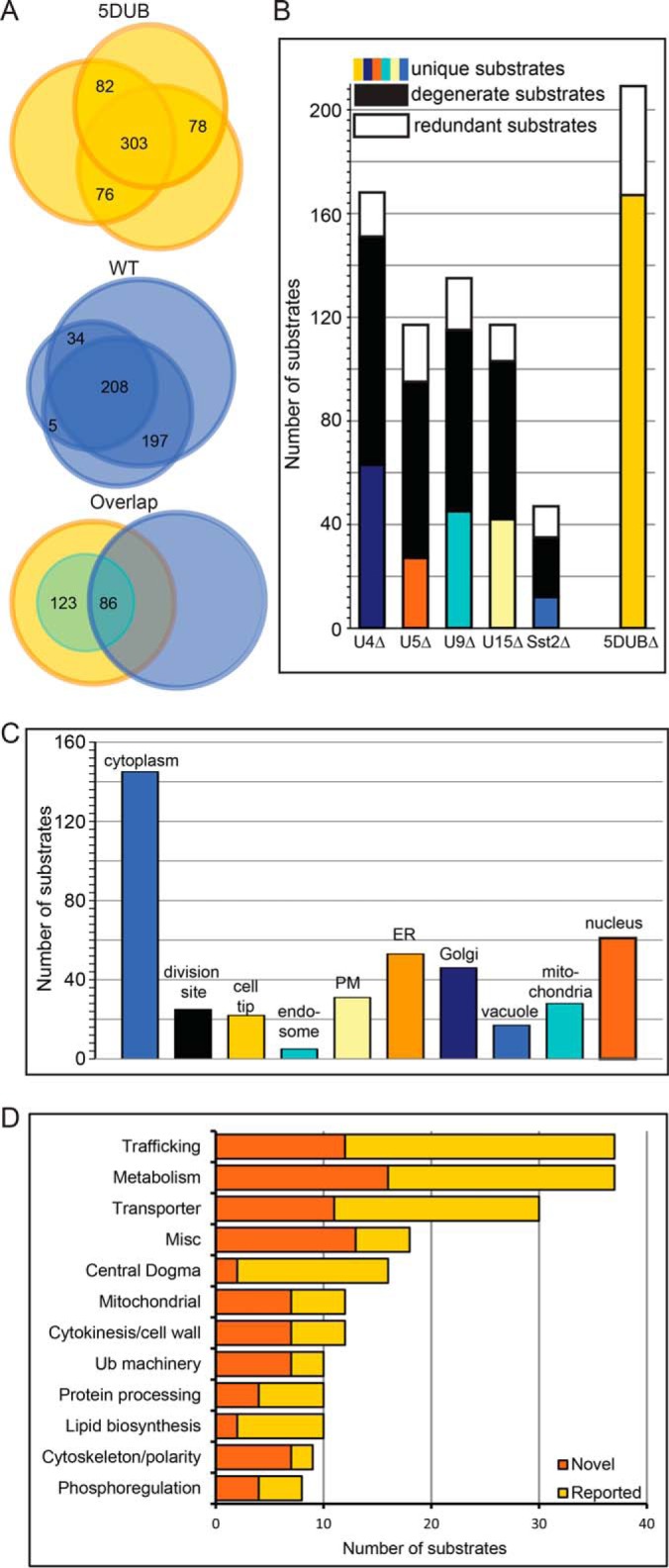
Comparative proteomics of wildtype (WT) and the 5DUB delete S. pombe strains reveals the degeneracy of these 5 DUBs. (A) Venn diagrams indicating the overlap of proteins identified in three independent experiments for the 5DUBΔ (upper) and WT (middle) strains. Bottom panel shows overlap of proteins identified in both strains and the small inset circle represents the putative substrates. (B) Bar graph showing numbers of substrates identified for each DUB using the indicated strains. Colored bars indicate substrates unique to specific strains, black bars indicate substrates shared with at least one other DUB, and white bars indicate substrates identified in at least one of the single deletions in addition to the 5DUB delete. (C) Cellular localization of the putative DUB substrates. Note that many substrates are present in multiple cellular locations. (D) Bar graph indicating the number of substrates for each gene ontology category (Pombase (50)) for the shared substrates of the five membrane trafficking DUBs. The dark orange bars represent ubiquitinated substrates that have not been reported as ubiquitinated, and the light orange bars represent substrates whose budding yeast orthologs have been reported as ubiquitinated proteins (17, 34, 35) (see Supplemental Table S6).
